43
CHAPTER III
44
1. Sera samples
A swine serum sample positive for both TTSuVs by conventional PCR (Segalés et
al., 2009), and a human serum sample TTV positive by real-time quantitative PCR (Maggi et al, 2005) were selected to optimize the Anello-RCA technique. After technical optimization, 10 swine and 5 human sera negative by the conventional PCR (Segalés et al., 2009) and real-time quantitative PCR (Maggi et al., 2005), respectively, were tested.
Furthermore, 159 serum samples from animal species including chicken (49),
bovine (68) and sheep (6), whose TTVs sequences have not been fully characterized as yet, and also several species of birds (36), whose at present anelloviral sequences have not been detected, were tested.
2. Samples purification and DNA extraction
Serum samples (250 µl) were filtered through 0.22 µm Ultrafree-MC sterile Centrifugal filter units (Millipore, Ireland) by centrifugation at 12 000 x g for 20 min at 25°C. To remove free nucleic acids, serum samples were treated with 50 U Benzonase Nuclease (purity > 99%, Novagen, Germany) at 37°C for 30 min, as previously described (Biagini et al., 2007). For evaluating the efficiencies of the filtration process combined with the enzymatic treatment, PCRs to test the presence of endogenous swine gene in pig serum were performed. One µl DNA (~ 30 ng) obtained concentrating the extraction products from a TTSuV1 and TTSuV2 positive pig serum was added to the reaction (25µl mix) containing 0.2 mM of dNTPs (Roche Diagnostic GmbH, Germany), 0.5 µM of forward and reverse primers specific for
45 endogenous swine gene [primers for: βLactoglobulin gene, βLG-F and βLG-R primers; Glucagon gene, GLC-F and GLC-R primers; Phospholipase C, pLCb-1-F and pLCb-1-R primers], 1x Green GoTaq Flexi Buffer (Promega, Madison WI U.S.A), 1.5 mM of MgCl2 (Promega, Madison WI USA) and 0.6 U of GoTaq Flexi (Promega,
Madison WI USA). The amplifications were initiated by heating at 94°C for 5 min followed by 35 cycles at 94°C for 15 sec, melting Temperature (Tm) [62°C; 62°C; 60°C, respectively] for 20 sec, and 72°C for 30 sec, and with final extension at 72°C for 5 min. Twenty µl of amplified product were controlled on 2% agarose gel. Thereafter, viral DNA was extracted using the NucleoSpin Blood kit (Macherey Nagel GmbH & Co. KG, Germany) for the serum samples from animal species and the QIAamp DNA Mini kit (Qiagen, Chatsworth, CA) for the human sera, resuspended in 50 µl of elution buffer, and stored at -20°C until use.
3. RCA- sequence-independent single primer amplification (SISPA)
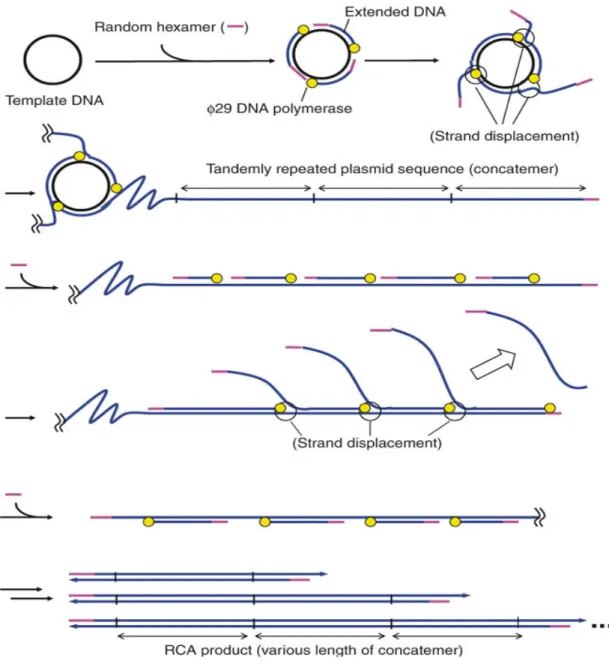
One µl DNA (~ 20 ng) obtained concentrating the extraction products from a TTSuV1 and TTSuV2 positive pig serum sample was denatured at 94°C for 3 min, cooled on ice and added to a reaction mixture containing 25 µM of random exo-resistant hexanucleotides (Fermentas, EU), 4mM dNTPs (Roche Diagnostic GmbH, Germany), 10U of phi29 DNA polymerase and its corresponding buffer (Fermentas, EU) in a final volume of 20 µl. Amplification was carried out at 30°C for 17 h, followed by inactivation of phi29 DNA polymerase at 65°C for 10 min. The resulting linear double-stranded DNA product (20 µl, ~ 2,5-3 µg/µl) (Fig. 5) was purified and concentrated to get a final volume of 10-15 µl (~ 4 µg/µl DNA) using 0.22 µm
46 Microcon Centrifugal filter devices Ultracel YM-100 (Millipore Corporation, Billerica).
For the SISPA procedure, 11 µg of the RCA product was digested for 3 h with 60 U of TaqI at 65°C (Fermentas, EU) and 60 U of Csp6I at 37°C (Fermentas, EU), enzymes that cut several times all TTV isolates. Digestion products were run on 1.3% agarose gel and fragments between 300 and 3000 base pairs were isolated from the gel using the NucleoSpin Extract II kit (Macherey Nagel GmbH & Co. KG, Germany). Subsequently, the isolated DNA fragments were ligated to the corresponding adaptors NTaq [200 µM hybridized oligonucleotides NTaq24, AGG CAA CTG TGC TAT CCG AGG GAT; and NTaq11, PO4=-CGA TCC CTC GG] and NCsp [200 µM
hybridized oligonucleotides NBam24, AGG CAA CTG TGC TAT CCG AGG GAG; and NCsp11, PO4=-TAC TCC CTC GG] (Invitrogen) (Allander et al., 2001; Biagini et
al., 2007), in a final volume of 50 µl containing 3U of T4 DNA ligase (Roche Diagnostic GmbH, Germany) and its corresponding buffer. Ligation was performed at 4°C for 16 h followed by enzyme inactivation at 70°C for 10 min. The ligation reactions were purified using 0.22 µm Microcon Centrifugal filter devices Ultracel YM-100 (Millipore Corporation, Billerica) and PCR amplified using the appropriate primers (0.4 µM NTaq24 or NBam24) (Allander et al., 2001; Biagini et al., 2007). The 50 µl PCR reactions contained 20-25 ng of DNA template, 5 U of La Taq Takara polymerase (TaKaRa LA Taq, TakaRa Bio Inc, Japan) with its corresponding 10 x GC II Buffer, and 2.5 mM of dNTPs (TaKaRa LA Taq, TakaRa Bio Inc, Japan). The polymerase was added into the mixture after the initial heating at 72°C for 3 min, followed by 5 min at 72°C. Amplification was initiated by heating at 95°C for 3 min,
47 followed by 35 cycles at 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min, and with final extension at 72°C for 30 min. Fifty µl of amplified product were run on 1.5% agarose gel and expected fragments were isolated from the gel using the NucleoSpin Extract II kit (Macherey Nagel GmbH & Co. KG, Germany).
48
4. Human TTV Detection and Quantification
Presence and load of TTV in human sera were determined by a universal TaqMan real-time PCR assay as previously described (Maggi et al., 2003a, 2003b) that was targeted to a highly conserved segment of the noncoding region of the viral genome. The procedures used for quantification of copy numbers and evaluation of specificity, sensitivity, intra and interassay precision, and reproducibility of the assay have been described (Maggi et al., 2003a, 2003b). TTV loads greater than 2 log10 DNA
copies/mL of serum were considered positive.
5. Anello specific RCA and PCR
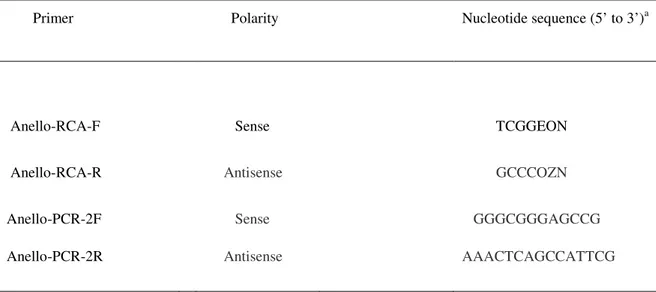
Based on the highly conserved region located near to the TATA box among several full-length genomes belonging to the family Anelloviridae from all available host species in the GenBank database (Okamoto, 2009b, Ng et al., 2009) (Fig. 6), short 7 bp exo-resistant, specific primers were designed (Table 3). The Anello-RCA was performed as described for the standard Anello-RCA, except using 12.5 µM of Anello-RCA-F and Anello-RCA-R primers. The resulting linear double-stranded DNA product was purified and concentrated as explained above. The purified Anello-RCA product diluted 1:100 (10-100 ng DNA) from swine sera was used as template in inverse-PCR by Anello-specific PCR primers (Table 3). For optimization in swine, different Tm (10 temperatures between 33°C and 60°C) and MgCl2 concentrations
(1.5, 2 and 2.5 mM) were tested. The optimal reaction (25 µl mix) contained 0.4 mM of dNTPs (Roche Diagnostic GmbH, Germany), 0.4 µM of Anello-PCR-2F and Anello-PCR-2R primers, 1x Green GoTaq Flexi Buffer (Promega, Madison WI
49 U.S.A), 1.5 mM of MgCl2 (Promega, Madison WI USA) and 0.75 U of GoTaq Flexi
(Promega, Madison WI USA). The amplification was initiated by heating at 94°C for 5 min followed by 45 cycles at 94°C for 1 min, 54.5°C for 1 min, and 72°C for 3 min, and with final extension at 72°C for 15 min. Twenty µl of amplified product were run on 1% agarose gel and fragments between 2 and 3 Kb were isolated using the NucleoSpin Extract II kit (Macherey Nagel GmbH & Co. KG, Germany).
Human sera were treated as explained above, and after different Tm (12 temperatures between 40°C and 60°C) and MgCl2 concentrations(1.5, 2 and 2.5 mM),
the optimized Anello-PCR reactions contained La Taq Takara (2.5 U) (TaKaRa LA Taq, TakaRa Bio Inc, Japan) with its corresponding 10 x GC II Buffer and 2.5 mM dNTPs. The Anello-RCA product diluted 1:1000 (~ 20 ng DNA) was used as template. Two different Tm were tested (40°C and 50°C). The amplification was initiated by heating at 94°C for 3 min, followed by 40 cycles at 94°C for 1 min, Tm for 1 min and 72°C for 4 min, with a final extension for 15 min at 72°C. Amplification products were run on 1% agarose gel and fragments between 3 and 4 Kb were isolated using NucleoSpin Extract II kit (Macherey Nagel GmbH & Co. KG, Germany).
Furthermore, pool of 3-4 serum samples from the 159 sera from the other species tested, including chicken, bovine, sheep and birds, were treated using for Anello-RCA/PCR the same conditions optimized for swine. Then, 36 out of 159 sera were used in Anello-PCR testing the conditions previously optimized for human. Amplification products were run on 1% agarose gel and each fragment observed was isolated as explained above.
50
Primer Polarity Nucleotide sequence (5’ to 3’)a
Anello-RCA-F Sense TCGGEON
Anello-RCA-R Antisense GCCCOZN
Anello-PCR-2F Sense GGGCGGGAGCCG
Anello-PCR-2R Antisense AAACTCAGCCATTCG
a
E denotes G, O denotes C, Z denotes T, N denotes any nucleotide. E, O and Z contain phosphorotioate groups.
Table 3. Nucleotide sequences of the specific primers used for Anello-RCA and
52
Fig. 6 Alignment of the region downstream the TATA box of full-length genomes belonging to the family Anelloviridae from several species available in the GenBank.
The position and polarity of Anello-RCA primers and Anello-PCR primers are indicated. Points indicate nucleotide identities between sequences, while dashes represent insertions/deletions. A stretch of 60 bp located between the primers Anello-PCR 2R and Anello-Anello-PCR 2F has been removed from the Figure and represented by a vertical zig-zag line.
6. Cloning
Gel isolated PCR products from RCA-SISPA and Anello-PCR were cloned into TOPO TA cloning vector according to the manufacturer’s instructions (Invitrogen, Carlsbad, USA). Single colonies were chosen for sequencing.
7. Sequencing
The cloned amplification products were sequenced using the BigDye Terminator
Cycle Sequencing Kit v.3.1 and ABI PRISM-3100 Genetic Analyzer (Applied Biosystems, Singapore). Twenty ng of each DNA template were sequenced in a 10 µl mix containing 1 µl BigDye Reaction Mix, 1x BigDye Sequencing Buffer and 10 µM M13-F and M13-R vector-specific primers (Invitrogen, Carlsbad, USA). The resulting sequences were assembled using the ContigExpress application (Vector NTI v11, Invitrogen) and analyzed by the BLAST program.
53
8. TTSuVs quantification
Quantitative PCR was used to estimate the TTSuV1 and TTSuV2 genome copies before and after Anello-RCA and RCA (Aramouni et al., 2010). TTSuV1 forward primer (TTSuV1F), TTSuV1 reverse primer (TTSuV1R), TTSuV2 forward primer (TTSuV2F) and TTSuV2 reverse primer (TTSuV2R) were designed using D-LUX™ Designer Desktop v.3.0 from Invitrogen and were predicted to work under universal conditions. TTSuV1F and TTSuV2R primers were labelled at the 3’ with JOE ™ (6-carboxy-dichloro-dimethoxy-fluorescein) and FAM ™ (6-carboxy-fluorescein), respectively. Amplicon sizes of TTSuV1 and TTSuV2 were 86 bp and 67 bp, respectively.
All primers were tested for cross-specificity to both TTSuV species, swine genome, PK-15 cell line DNA, and the most common swine viruses like PRRSV, PPV, PCV1, and PCV2 genotypes “a” (PCV2a) and ”b” (PCV2b), by using the BLAST software and in direct qPCR assays.
For the standard preparations, TTSuV1 and TTSuV2 full-length genomes were amplified with proof reading activity polymerase (TaKaRa LA Taq ™) and specific pairs of primers (TTSuV1: sense: 5' TGA GTT TAT GCC GCC AGC GGT AGA 3’; antisense: 5' GCC ATT CGG AAC TGC ACT TAC T 3’; TTSuV2: sense: 5' GAA TTC GCT AGA TTT TTA AAA GGA AAG 3’; antisense: 5' GAA TTC CAT TCC AAC ATT ACT AGC T G 3') and then cloned into the pCR2.1 vector. Plasmid purifications were made using the Qiaprep Spin Miniprep kit (Qiagen, Valencia, CA, USA) according to the manufacturer instructions. After a spectrophotometric quantification of the plasmids, standards were prepared in serial dilutions ranging
54 from 109 to 101 molecules/µl. Two µl of the standards ranging between 105 and 10 molecules/µl were used subsequently for the quantification of TTSuV1 and TTSuV2 in the studied samples.
Reactions were carried out in 96-well plates. Each sample and standards were run in triplicate and a negative control was added between each three wells, using autoclaved distilled water (Sigma-Aldrich, UK). Each reaction contained 2 µl of sample or standard DNA, 200 nM of each primer (TTSuV1: sense: 5' CGA CCG GAG TCA AAT CTG ATT GGT [JOE] G 3’; antisense: 5' TAC TGG GAA CGC CCT AAT TCT G 3’; TTSuV2: sense: 5' CGG TTG AAC AGA GCT GAG TGT CTA AC [FAM] G 3’; antisense: 5' CCC TTG ACT CCG CTC TCA GG 3'), 10 µl of Express qPCR Supermix Universal (Invitrogen, Carlsbad, USA) and 0.04 µl of Rox in a final volume of 20 µl. Amplification and quantification were perfomed using the ABI®7500 Fast Real Time PCR System (Applied Biosystems, Singapore) under universal conditions: 10 min at 95°C, 2 min at 50°C and 40 cycles of 15 s at 95°C, 1 min at 60°C. TTSuV loads greater than 3,69 log10 DNA copies/mL of serum were considered positive
samples. Typical Amplification Plot and its corresponding Standard Curve are shown in figure 7.
55
Fig. 7 Typical Amplification Plot and corresponding Standard Curve in real-time