Selective influences in the expressed immunoglobulin
heavy and light chain gene repertoire in hairy cell
leukemia
Francesco Forconi,
1Elisa Sozzi,
1Davide Rossi,
2Surinder S. Sahota,
3Teresa Amato,
4Donatella Raspadori,
1Livio Trentin,
5Lorenzo Leoncini,
2Gianluca Gaidano,
2and
Francesco Lauria
11
Sezione Ematologia e Trapianti, Università di Siena; 2Divisione di Ematologia, Dipartimento di Medicina Clinica e
Sperimentale and Centro Interdisciplinare di Biotecnologie per la Ricerca Medica Applicata, Università del Piemonte
Orientale Amedeo Avogadro, Novara, Italy; 3Cancer Sciences Division, University of Southampton, UK; 4Sezione di
Anatomia Patologica, Università di Siena, and 5Dipartimento di Medicina Clinica e Sperimentale, Divisione di
Ematologia-Immunologia, Università di Padova, Italy
Funding: the work was supported by an EHA clinical research grant 2004, and grants from the Hairy Cell Leukemia Foundation (Illinois, USA), Piano di Ateneo per la Ricerca (University of Siena),
PRIN-MUR 2006 (Progetto di Ricerca di Interesse Nazionale - Ministry of University and Research), Ricerca Sanitaria Finalizzata, Regione Piemonte, Novara-AIL Onlus, Associazione Italiana per la Ricerca sul Cancro (AIRC, Italy), Siena-AIL Onlus. SSS is supported by the Leukaemia Research Fund UK. Manuscript received September 24, 2007. Revised version arrived on November 22, 2007. Manuscript accepted December 27, 2007. Correspondence:
Francesco Forconi, MD, DM, PhD, Sezione di Ematologia e Trapianti, Dipartimento di Medicina Clinica e Scienze Immunologiche, Università di Siena, AOUS, viale Bracci, 53100 Siena, Italy E-mail: [email protected]
The online version of this article con-tains a supplementary appendix.
ABSTRACT
Background
We previously reported ongoing mutational and isotype switch events in the immunoglobulin (Ig) heavy chain (H) locus in hairy cell leukemia. Those analyses raised questions on the incidence and type of selective influences occurring on the tumor B-cell receptor of hairy cell leukemia.
Design and Methods
To further investigate this issue, we examined the full IGH and k and l light chains (IGk and IGl) variable and constant region transcripts expressed in a large cohort of patients with hairy cell leukemia (n=88).
Results
Multiple IgH isotypes were expressed in 46/56 (82%) cases of hairy cell leukemia. Comparison of tumor with normal B-cell repertoires revealed preferential usage of IGHV3-21, IGHV3-30 and IGHV3-33 in hairy cell leukemia (p=0.001, p=0.003 and p=0.001, respectively). Light chain analysis demonstrated preferential Igl use with an inverted IGk:IGl ratio (0.7:1) and universal usage of IGLJ3. Analysis of LCDR3 junctions revealed highly homologous motifs in 40% of IGL. Parallel analysis of IGH and IGL showed selective pairing of IGHV3-21/30/33 segments to spe-cific LCDR3-J3 subsets (p=0.008). Of 40 cases of hairy cell leukemia, 38 had mutated IGHV and/or IGK/LV, with variations in 13/13 cloned cases, while two had 100% unmutated IGHV and IGK/LV.
Conclusions
Overall, biased IGV usage, preference for Igl with universal IGLJ3 usage and a high incidence of LCDR3 homologous motifs suggest selective influences on the B-cell receptor of hairy cell leukemia. Ongoing mutations and isotype switching suggest that influences occur on the tumor B-cell receptor at ectopic sites.
Key words: hairy cell leukemia, immunoglobulin, IGH, IGL, IGK, heavy chain, light chain. Citation: Forconi F, Sozzi E, Rossi D, Sahota SS, Amato T, Raspadori D, Trentin L, Leoncini L, Gaidano G, and Lauria F. Selective influences in the expressed immunoglobulin heavy and light chain gene repertoire in hairy cell leukemia. Haematologica 2008 May; 93(5):697-705. doi: 10.3324/haematol.12282
Introduction
Hairy cell leukemia (HCL) is a rare, chronic B-cell neoplasm characterized by leukemic hairy cells present in blood, bone marrow, and splenic red pulp, with atro-phy of white pulp. Lymph node involvement is infre-quent. HCL is typically associated with markers of acti-vation, which include expression of CD25, CD11c, FMC7, and CD103 at high intensity.1A distinctive
fea-ture of HCL is expression of multiple surface (s) immu-noglobulin (Ig) isotypes, although their prevalence in HCL has not been fully mapped.1,2
B-cell tumors preserve the B-cell receptor (BCR) fea-tures of the originally transformed cell.3Consequently,
immunoglobulin gene (IG) analysis delineates the criti-cal events of clonal development and defines the IG heavy (H) and light κ (K) or λ (L) repertoire selected by specific tumor entities.3In some instances, IG analysis
may also have prognostic value,3,4 and the selected
IGH/IGL or IGH/IGK pairs can associate with specific
BCR structure and clinical behavior.4Selective stimuli
to the tumor BCR may be of different types, including viral or bacterial antigens, or, in germinal center-derived lymphomas, stromal elements acting on N-glycosylat-ed residues acquirN-glycosylat-ed by somatic mutation.5,6Analysis of
the selective influences on the tumor BCR has often been hampered in HCL by the rarity of the disease, and only small series of cases have been analyzed to date.1
In small series of HCL, we and others have observed that most HCL carry mutated IGH variable region (V) genes, with low levels of intraclonal heterogeneity. Only a minor subset of HCL has unmutated IGHV genes.2,7-9Both mutated and unmutated subsets of HCL
express multiple sIgH isotypes with no evidence of subpopulations.2 Also, activation-induced
cytidine-deaminase is expressed in HCL, and Ig sterile tran-scripts are produced prior to class switch deletional recombination.2However, hairy cells fail to express the
germinal center markers CD38, CD10 and BCL6, or the memory B-cell marker CD27.10,11 Most importantly,
studies of gene expression profiling of B cells from dis-crete normal subsets or HCL have shown that hairy cells are related to memory cells, although with altered expression of genes controlling cell adhesion and chemokine-receptors.10 This raises questions on the
incidence of the BCR events and where they occur in HCL.
The observation that HCL biology may not be sim-ply recapitulated by normal B-cell physiology raises several unresolved issues. These include the signifi-cance of ongoing somatic mutation and isotype switch events in HCL, and the role of selective influences in shaping the BCR repertoire of HCL. Here we addressed these issues by investigating the Ig heavy and light chains expressed by the tumor cells of a large series of patients with HCL.
Design and Methods
Patients
Peripheral blood samples were collected from 88 patients with HCL. In all instances, the specimens were collected at diagnosis before specific therapy. The diag-nosis of typical HCL and variant HCL was based on peripheral blood morphology, flow cytometry and bone marrow immunohistochemistry according to the World Health Organization classification.12 Informed
consent was obtained in all cases and the study was approved by the local Institutional Review Board.
Phenotypic analysis
Peripheral blood mononuclear cells were obtained by Lymphoprep (Nycomed Pharma, Oslo, Norway) gradi-ent separation. Immunophenotypic studies were car-ried out on these cells by direct immunofluorescence techniques with a large panel of antibodies.7Expression
of CD27, CD38 and sIgH isotypes on HCL was deter-mined by three-color staining with F(ab’)2 anti-sIgG, anti-sIgM, anti-sIgD, and anti-sIgA antibodies.7
Expression of sIgκ/λ was determined by three color staining with fluorescein isothiocyanate (FITC)–conju-gated F(ab’)2 anti-sIgλ, phycoerythrin (PE)-conju(FITC)–conju-gated F(ab’)2 anti-sIgκ and peridinin-chlorophyll protein (PerCP)-conjugated anti-CD20. Previously described procedures were used to avoid non-specific binding.7
Data were acquired and analyzed and antigen expres-sion was defined as described elsewhere.7
Polymerase chian reaction amplification of IGHVDJ,
IGKVJ and IGLVJ transcripts and sequence analysis
Total RNA was isolated from peripheral blood mononuclear cells and cDNA prepared as described previously.7 The full tumor IGHVDJ transcripts wereamplified by PCR with a mixture of Leader-VH-mix primers and a constant-region primer.7The full tumor
IGKVDJ or IGLVJ transcripts were identified by PCR
using a mixture of 5’ primers specific for known IGK or
IGL leader sequences (IgKV- or IgLV-leader mix),
together with a 3’ primer specific for either the IGK or
IGL constant-region.13Amplified products were run on
agarose gel and purified with a Jet Quick Gel extraction kit (Celbio, Milan, Italy). The tumor IGHVDJ, IGKVJ and IGLVJ sequences were identified by direct sequenc-ing and/or after clonsequenc-ing the purified band. Ligation and cloning were performed with pGEM®-T Easy Vector
System II and JM109 competent cells (Promega, Milan, Italy). Sequencing was carried out using the v1.1 Big Dye Terminator Ready Reaction sequencing kit (AB Applied Biosystems, Applera Italia, Monza, Italy), on an ABI Prism 310 genetic analyzer (PerkinElmer, Warrington, UK). Direct sequencing was performed with the 3’ primer on the constant region and the iden-tified sequence was confirmed with the family specific leader 5’ primer, which allowed identification of the full V-gene transcript. When cloning was performed, M13 forward and reverse primers were used to
sequence in both directions. The data were analyzed using Chromas 1.51 software and aligned to the 2005 updated V-BASE and ImMunoGeneTics (IMGT) data-bases.14,15
IGHV, IGKV and IGLV gene usage and mutation
pat-terns were analyzed as previously described.7 IMGT
nomenclature was used to assign IG gene use,14since
this allowed comparison with data from previously used nomenclatures for IGH, IGK and IGL genes.16,17
Lengths in the IGH and IGK/LCDR3 (HCDR3, KCDR3 and LCDR3, respectively) were calculated according to IMGT criteria.14IMGT criteria and nomenclature were
also used for IGHD determination, again allowing com-parison to segments with other designations from the literature.14N-addition of G and C at the joining ends of
the V(D)J junction [(Ngc) Online Supplementary Table S1] was performed to investigate TdT activity. Recurrent amino acid sequence motifs in HCDR3, KCDR3 and LCDR3 were sought using the ClustalW tool (at
http://www.ebi.ac.uk/clustalw/#). Amino acid identity
>60% in the HCDR3 or >80% in the K/LCDR3 was required for the inclusion of an IgH or an IgK/L chain in the same subset, respectively.18 Intraclonal
heterogene-ity was assessed in the cloned products and was distin-guished from Taq infidelity by an increased frequency compared to Taq error rate and by the finding of the same mutation in more than one clone.7If only direct
sequencing was performed, the tumor IGHV, IGKV and IGLV sequences were confirmed by replicate RT-PCR and sequencing. The incidence of potential novel N-glycosylation sites in IGHVDJ, IGKVJ and IGLVJ transcript sequences was assessed as previously described.6,19 Statistical analyses were performed using
χ2or Fisher’s exact tests. p values <0.05 were
consid-ered statistically significant.
Results
Analysis of expressed IGHV region transcripts and
sIgH isotype proteins
The expressed tumor IGHV sequences were identi-fied in 83/88 HCL (Online Supplementary Table S1). The distribution of IGHV families was similar to that of normal B-cells, but use of individual IGHV gene seg-ments showed differences from that of the normal B-cell repertoire (Figure 1A). The IGHV gene segments most frequently used in HCL were IGHV3-30 (13/83, 16%), IGHV3-33 (9/83, 11%), IGHV3-23 (7/83, 8.5%),
IGHV3-21 (7/83, 8.5%), IGHV4-30/4-31 (5/83, 6%)
and IGHV4-34 (5/83, 6%). Among these segments, the usage of IGHV3-21, IGHV3-30 and IGHV3-33 was sig-nificantly increased compared to the normal B-cell repertoire (p=0.001, p=0.003 and p=0.001, respective-ly).20Other IGHV segments were used less frequently
in HCL than in normal B cells (Figure 1A), although the differences were not statistically significant, for any of the segments individually, likely due to the number of cases investigated. However, by combining results of this study with those of all published HCL IGHV gene sequences (n=164, Online Supplementary Figure S1A), we confirmed the overuse of IGHV3-21, IGHV3-30 and
IGHV3-33, and demonstrated the significantly reduced
use of IGHV1-18 (p=0.02) and IGHV3-53 (p=0.02).8,9,21,22
Phenotypic analysis of surface IgM, IgD, IgG and IgA was performed in 56 HCL (Online Supplementary Table
S1). The majority (46/56, 82%) co-expressed multiple
pre- (IgM±D) and post-switch (IgG±A) isotypes, indi-cating that multiple isotype expression is a dominant feature in HCL.2Of 63 HCL tested, CD38 was negative
in all cases, and the memory B-cell marker CD27 was negative in 58/63 cases. Interestingly the five CD27-positive cases (cases HCL8, HCL17, HCL72, HCL82, and HCL87) differed from all the remaining HCL inves-tigated, as they had features consistent with the diag-nosis of variant HCL, including high white blood cell counts with prolymphocytic morphology and CD25 Figure 1. Expressed IGHV, IGHK and IGLV region repertoire in HCL.
The incidence of IGHV (A), IGKV (B) and IGLV (C) region transcripts expressed by HCL was compared to the repertoire of normal B cells from published data.20,24,25
Dark gray columns: HCL; light gray columns: normal B-cell repertoire. Asterisks indicate V genes whose incidence was a significantly different between HCL and the normal B-cell repertoire.
A HCL Normal 1-02 1-03 1-18 1-46 1-69 2-05 3-07 3-09 3-11 3-15 3-21 3-23 3-30 3-33 3-48 3-49 3-53 3-66 3-74 4-30/4-31 4-34 4-39 4-59 4-61 4b 5-51 5a 6-01 1-17 1-5 1-9 1D-17 1D33 1D-37 1-D39 3-11 3-15 3-20 4-1 6D-21 1-16 1-40 1-44 1-47 1-51 2-11 2-14 2-8 3-21 4-69 6-57 7-43 9-49 16 14 12 10 % 8 6 4 2 0 IGHV 30 25 20 % 15 10 5 0 IGHV 20 18 16 14 12 % 10 8 6 4 2 0 IGLV B C
negativity of the peripheral blood hairy cells.12These
data confirm that lack of surface CD27 and CD38 is a feature of typical HCL.11
Analysis of HCDR3 junction
Sixty-nine cases of HCL were evaluable for the
HCDR3 junction (Online Supplementary Table S1 and
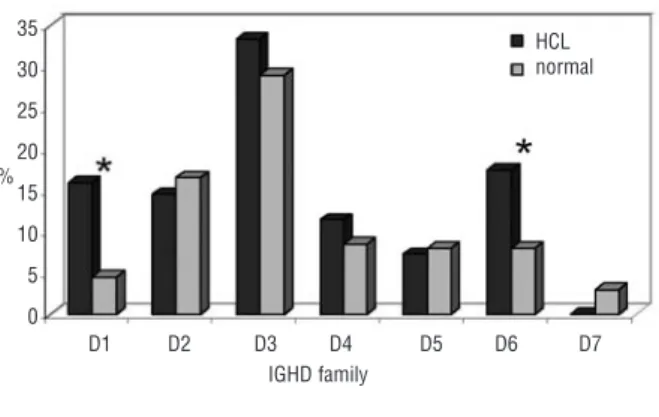
Figure 2). IGHD gene segment analysis revealed signif-icantly increased use of IGHD1 (11/69 cases, 16%) and
IGHD6 (12/69, 17%) families (p=0.001 and 0.02,
respectively). Analysis of specific segments revealed additional notable biases. In fact, the IGHD1 family used almost exclusively IGHD1-26 (10/11 cases) and the IGHD6 family frequently utilized IGHD6-19 (5/12 cases). This observation is remarkable, since neither of these segments has been reported to be used in normal B cells.20In addition, significantly increased selection of
IGHD3-3 (8/69, p=0.00009), IGHD3-9 (4/69, p=0.004), IGHD3-10 (7/69, p=0.00005) and IGHD4-17 (5/69, p=0.0009) was also documented. Again, IGHD3-3, IGHD3-9, and IGHD4-17 are not reportedly selected in
the functional normal B-cell repertoire, and only
IGHD3-10 is restricted to 0.5% of all D segments.20
IGHJ gene segments used in the expressed HCL
reper-toire distributed similarly to those in the normal B-cell repertoire (Online Supplementary Table S1), HCDR3 length was variable (range 9-28 amino acids, median 16, mean 17), and analysis of recurrent amino acid sequence motifs could not identify significant simili-tudes of HCDR3. However, the number of sequences investigated (n=69) here and the few HCDR3 currently available in the literature (n=27) preclude any statistical clustering.8,22,23 Thus, the significance of IGHD biases
remains unresolved as yet.
Analysis of sIg
κ and sIgλ protein expression and
IGKV and IGLV region transcripts
Seventy of 83 HCL were characterized for sIgκ and sIgλ protein expression (Online Supplementary Table S1). Expression of sIgλ was observed in 41/70 (59%) HCL with an Igκ:Igλ ratio of 0.7:1, indicating preferential secondary rearrangement of Igλ. The expressed IGK and IGL tumor transcript sequences were investigated in 45/70 cases (23 HCL expressing sIgκ and 22 HCL expressing sIgλ). In two additional samples (HCL30 and HCL33), IGL transcripts from the non-functional allele were amplified and sequenced. Four HCL (HCL4, HCL42, HCL70, and HCL83) co-expressing double IGK or double IGL, or IGK/IGL, were excluded from the analysis. Among IGKV gene segments (Online
Supplementary Table S2 and Figure 1B), IGKV1D-39 (012/02) was the one most frequently used (6/23, 26%),
and its usage was significantly higher than in normal B-cells (p=0.03).24 The distal gene segments IGKV1D-17
(case HCL63) and IGKV6D-21 (case HCL11), which are not reported in the normal B-cell repertoire, were both used once. The other IGKV segments distributed simi-larly to normal B-cells.
Among functional IGLV gene segments, IGLV2-14 was most frequently used (4/22, 18%), followed by
IGLV1-47 (3/22, 14%), IGLV1-40, IGLV1-44, IGLV1-51, IGLV2-11 and IGLV3-21 (2/22, 9% each) (Online
Supplementary Table S2 and Figure 1C), with an overall
distribution similar to that of the normal B-cell reper-toire.25
Analysis of KCDR3 and LCDR3 junctions
Twenty-one HCL were evaluable for the KCDR3 junction. IGKJ genes were used by HCL in a fashion similar to that in the normal B-cell repertoire (Online
Supplementary Table S2).24KCDR3 length ranged from 7
to 11 amino acids (median 9) and 11/21 KCDR3 had identical pI (range 6.5-13, median 13). Overall analysis of KCDR3 amino acid sequences reflected pI similari-ties and identified three subsets, all with 88.8% sequence identity (Online Supplementary Table S2). Subset 1K (HCL28 and HCL2) harbored
IGKV1D-33-J1/4 rearrangements (QQYDNLP[L/R]T), which
associ-ated with IGHV3-23 (HCL28) or IGHV1-02 (HCL2). Subset 2K (HCL7/330 and HCL67) harbored
IGKV3-20-J1/2 rearrangements (QQYGRSP[Q/Y]T), which
associated with IGHV2-05 (HCL7/330) or IGHV4-34 (HCL67). Subset 3K (HCL19 and HCL63) harbored
IGKV1-17/1D-17-J1/2 rearrangements
(LQHNSYP[R/Y]T), which associated with IgHV3-21 (HCL19) or IgHV4-34 (HCL63).
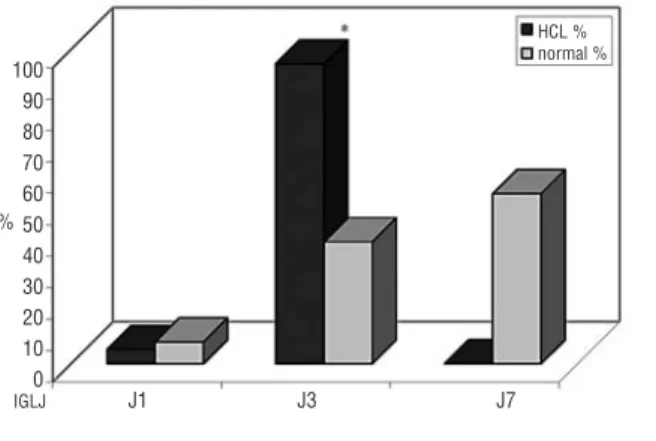
Twenty-two HCL were evaluable for the functional LCDR3 junction. Remarkably, among IGLJ segments, virtually all cases used IGLJ3 (21/22, 95.5%) (Online
Supplementary Table S2 and Figure 3). The universal use
of IGLJ3 was higher than expected by chance alone (50%), and significantly higher than its frequency (34%) in normal B cells (p=0.000000001).26Remarkably,
2/2 HCL with non-functional LCDR3 junctions (HCL30 and HCL33) failed to use IGLJ3, further indicating strik-ingly selective influences on the functional repertoire. In particular, HCL30 used IGLJ1 with a TAG stop at joining codon 115, whereas HCL33 used IGLJ6 with and out-of-frame rearrangement in codon 115 (Online
Supplementary Table S2). LCDR3 length ranged from 9
to 13 amino acids (median 11) and the pI ranged from 4.4 to 13 (median 13), with 13/22 LCDR3 having an Figure 2. Expressed IGHD families in HCL. Incidences of IGD fam-ilies expressed by HCL were compared to the repertoire of normal B-cells from published data.20Dark gray columns: HCL; light gray columns: normal B-cell repertoire. Asterisks indicate regions with identified significant differences between HCL and the normal B-cell repertoire. The D1 family was represented by D1-26 in 10/11 cases. The D6 family was represented by D6-19 in 5/12 cases. D1-26 and D6-19 are not reportedly used by normal B cells.20
D1 D2 D3 D4 D5 D6 D7 IGHD family % 35 30 25 20 15 10 5 0 HCL normal
identical pI.
Analysis of LCDR3 amino acid sequences identified three HCL subsets (Figure 4). Subset 1L (HCL35, HCL22, HCL44, HCL49, and HCL40) harbored
IGLV1-44/47-J3 rearrangements with ≥85% homologous
LCDR3 (Figure 4). Of note, these HCL paired only with the highly identical IGH3-30 or IGHV3-33 segments in four out of five cases, the exception being case HCL22, which used IGHV3-21. Remarkably, the selective clus-tering of IGHV3-21/30/33 segments (5/5, 100%) within this subset was significant, when compared to pairing of IGHV3-21/30/33 segments with the remaining CDR3 investigated for IGLV-J (8/17, 47%, p=0.03) or the total IGL/KV-J (13/38, 34%, p=0.005) rearrange-ments (Online Supplementary Table S2). Subset 2L (HCL27 and HCL32) harbored IGLV3-21-J3 rearrange-ments with 91% homologous LCDR3 (QVWDSSS-DH[W/V]V), and associated with either IGHV3-30 or
IGHV3-23. Subset 3L (HCL64 and HCL23) harbored IGLV1-40-J3 rearrangements with 81.8% homologous
LCD3 (QSYD[S/N]SL[SG/TR]SGV), and associated with IGHV4-34 or IGHV3-21. Overall, the overused
IGHV3-30, IGHV3-33 and IGHV-3-21 genes clustered
within the identified LCDR3 sets (7/9 cases; 78%), and not with the CDR3 from the remaining IGK/LV-J rearrangements investigated (9/34, 26%, p=0.008). Together with Igλ light chain preference and universal
IGLJ3 use, these data further suggest that selection
events are dominant in the Igλ light chain of HCL. TdT activity was absent in almost 50% light chain rearrangements (20/41 light chain junctions). Since TdT activity decreases during B-cell ontogeny,27 lack of
N-addition/deletion of G and C at the ends of V-J junction (Online Supplementary Table S2) is suggestive of rear-rangements having occurred late (in the periphery) as a result of receptor revision in the light chain, after downregulation of the TdT enzyme.14
Somatic mutation analysis of IGHV, IGKV and IGLV
The mutation status of IGHV tumor transcripts was evaluated in 83 HCL (Online Supplementary Table S1). Overall, IGHV genes carried variable tiers of mutations (range 80.95-100%; median 96.56%, mean 95.97%). Three of 83 HCL had completely unmutated IGHV genes (100% homology to germline). However, by using the arbitrary 2% cut-off that defines mutational status in other lymphoproliferative disorders and that is being used for clinical correlates in ongoing clinical studies,28 the unmutated HCL subgroup expanded to
17/83 HCL (20.5%).
The distribution of individual IGHV genes varied among mutated and unmutated cases of HCL (Online
Supplementary Table S2 and Figure 5A). In particular, IGHV3-30, the segment most frequently used in HCL,
showed a selective tendency to distribute in unmutated rather than mutated HCL cases (6/17, 35% unmutated HCL vs. 7/66, 10% mutated HCL, p=0.01). IGHV4-34 was also more frequently used in unmutated HCL (3/17, 18%) than in mutated cases (2/66, 3%) (p=0.02).
IGKV mutation status (n=23) (Online Supplementary Table S2 and Figure 5B) was variable (homology range
93.61%-100%, median 97.26%, mean 97.31%), and
2/23 cases had completely unmutated IGKV genes. Using the arbitrary 2% cut-off value, 9/23 (39.1%)
IGKV were considered unmutated. IGLV mutation
sta-tus was evaluable in 22 cases with functional IGLV rearrangements (Online Supplementary Table S2 and Figure 5C). IGLV homology to germline ranged from 89.9% to 100% (median 97.12%, mean 96.83%), and 1/22 HCL had completely unmutated IGLV genes. Using the arbitrary 2% cut-off, 8/22 (36.4%) IGLV were unmutated.
Parallel assessment of IGHV and IGKV/IGLV muta-tion status was feasible in 40 cases of HCL (Online
Supplementary Table S2). The IGHV-IGKV/IGLV
muta-tion status was concordant in 22/40 HCL. Eighteen cases were mutated in both IGHV and IGKV/IGLV. Figure 3. IGLJ segment usage in IGλ HCL. The incidence of IgLJ
segments in the functional IGLVJ transcripts expressed by HCL was compared to the functional repertoire of normal B cells from published data.26
Dark gray columns: HCL; light gray columns: nor-mal B-cell repertoire. *IGLJ3 segment use in HCL was universal
(>95% cases) and significantly superior to IGLJ3 use in normal B cells.
Figure 4. Subsets of highly similar LCDR3 in HCL. Amino acid sequences are shown for all HCL cases within a LCDR3 subset. The rearranged IGLV and IGLJ and the associated IGHV gene seg-ment are indicated together with the amino acid sequence of each case. Amino acid homology to the first LCDR3 from each subset is indicated by dots. HCL belonging to these subsets pref-erentially utilized the most frequent IGHV3-21, 3-30 or 3-33 gene segments. (*, **, ***): *selective distribution of IGHV3-21, 3-30 or
3-33 in HCL from subset 1L compared to Igλ-positive HCL not
belonging to subset 1L; **selective distribution of IGHV3-21, 3-30 or 3-33 in HCL from subset 1L compared to Igλ-positive or Igκ-pos-itive HCL with light chain CDR3 not belonging to subset 1L; ***selective distribution of IGHV3-21, 3-30 or 3-33 in HCL from subsets 1L, 2L and 3L compared to HCL with light chain CDR3 not belonging to these subsets. ^An identical LCDR3 was reported in 2/368 normal or autoreactive B cells (0.5%), with no indication of antigen reactivity. °Identical LCDR3 in 5/368 normal or autoreac-tive B-cells (1.3%), that identified anti-platelet αIIbβ3 integrin
(EBA11) or anti-La (SS-B)SLE autoantibodies.46
HCL % normal % 100 90 80 70 60 50 40 30 20 10 0 % IGLJ J1 J3 J7 Subset 1L HCL35 1-44CAAWDASLNGV VF 3 HCL22 1-44...D....HY .. 1 HCL44/421-47...S.W .. 3 HCL49 1-47...S.G .. 3 HCL40v 1-47...R. .. 3 Subset 2L HCL27 3-21CQVWDSSSDHW VF^ 3 HCL32 3-21...V ..° 3 Subset 3L HCL64 1-40CQSYDSSLSGSG VF 3 HCL23 1-40...N....TR .. 3
IGLV LCDR3 IGLJ PairedIGHV
Probability of IGHV3-21, 3-30 or 3-33 associating with LCDR3 subsets
(*) (**) (***) 3-30 3-21 3-33 3-33 3-30 3-23 3-30 4-34 3-21 p∆=0.034 p∆=0.005 p∆=0.008
Four were unmutated in both IGHV and IGKV/IGLV. Of these, two cases displayed both IGHV and
IGKV/IGLV rearrangements with a 100% homology to
closest germline, confirming the existence of a very minor subset of HCL with completely unmutated IG genes.2
The IGHV-IGKV/IGLV mutation status appeared dis-cordant in 18 cases of HCL (12 IGHV
mutated-IGKV/IGLV unmutated cases and six IGHV
unmutat-ed-IGKV/IGLV mutated cases). However, 16/18 discor-dant cases carried some level of mutation (homology to germline <100%) in both IGHV and IGKV/IGLV, sug-gesting that the variations likely represent true muta-tions and not polymorphisms.29 Only two discordant
cases (HCL37 and HCL81) carried heavily mutated
IGHV (94.79% homology) or IGKV (94.98%) genes
coupled to completely unmutated (100%) IGKV or
IGHV genes, respectively. These two particular cases
might be representative of an antigen experienced BCR (with mutated IGH or IGK) rescued after secondary recombination of the IGK or IGH chain on the second allele, and suppression of the first functionally rearranged allele (receptor revision).30,31
Analysis of intraclonal heterogeneity of IGHV, IGKV
and IGLV
Cloning and sequencing of IGHV and IGKV or IGLV transcripts was performed in 12 cases of HCL (Table 1). In all cases, cloning confirmed the results of direct sequencing. Cloning of IGHV transcripts revealed intr-aclonal variations within the same or different tumor isotypes in 11/12 cases, including 2/2 cases with >98% and <100% IGHV homology to germline (cases HCL7 and HCL35). Cloning of the paired IGK/L tumor sequence confirmed intraclonal variations in the light chain of the same 11/12 cases. Using stringent criteria for ongoing activity (i.e. ≥2 identical variations repeat-ed in separate clones), intraclonal heterogeneity was restricted to 3/11 cases. Lack of repetitions in the remaining 8/11 cases may be due to the mutational fre-quencies in the light chain genes, which, in normal individuals, are generally lower than in the IGHV genes.32 Only HCL38, having both IGHV and IGLV
genes with 100% homology to germline, did not dis-play intraclonal mutations in either IGHV or IGLV genes.
Discussion
Our immunogenetic analysis of the expressed IGH and IGK/L repertoire in a large cohort of patients has implications for the origin of HCL, and indicates that
IG selection may play an important role in the
patho-genesis of this disease. IGHV analysis shows that HCL is characterized by biased usage of 21,
IGHV3-30 and IGHV3-33, in agreement with prior analyses on
smaller HCL groups.9,21IGHV3-30 and IGHV3-33 are
highly homologous segments with a single amino acid difference in the CDR2, are recognized by the same anti-CDR1 (B6) monoclonal antibody, and thus may share the ability to bind identical antigens.33 Indeed,
IgHV3 family members, including IGHV3-30 and
IGHV3-33, react with common bacterial superantigens,
such as modified staphylococcal protein A,34 or with
the natural staphylococcal enterotoxin A, which is suf-ficient to induce survival of IGHV3-expressing B cells by low-affinity binding.35 Furthermore, IGHV3-30 is
reportedly involved in the immune response against
Toxoplasma infection, providing additional clues to
potential antigens sustaining HCL.36Clearly, the
selec-tive influences acselec-tive in HCL appear to follow routes that are different from those of other B-cell neoplasms, in particular from the extensively investigated IgM-positive chronic lymphocytic leukemia (CLL).19 For
example, IGHV1-69 was totally absent in HCL from Figure 5. Distribution of mutated and unmutated IGHV (A), IGKV
(B) and IGLV (C) segments in HCL. Incidence of tumor IGHV, IGKV
and IGLV with homology to germline inferior (mutated) or superi-or (unmutated) to 98% in HCL. Segments with <98% homology are indicated by dark gray columns; segments with >98% homol-ogy to germline are indicated by light gray columns. Asterisks indi-cate segments with significantly different distribution between mutated and unmutated cases of HCL.
HCL<98% HCL>98% 40 35 30 25 20 15 10 5 0 IGHV 40 35 30 25 20 15 10 5 0 IGHV % % 30 25 20 15 10 5 0 IGLV % A B C 1-02 1-03 1-18 1-46 2-05 3-07 3-09 3-11 3-15 3-21 3-23 3-30 3-33 3-48 3-49 3-53 3-66 3-74 4-30/4-31 4-34 4-39 4-59 4-61 4b.01 5-51 5a 6-01 1-17(A30) 1-5(L12) 1-9(L8) 1D-17(L14) 1D-33(018/08) 1-D39(012/02) 3-11(L6) 3-15(L2) 3-20(A27) 4-1(B3) 6D-21(A26/A10) 1-16(L1) 2-28(A19/A3)
our and previously published series (total 164 IgHV sequences),2,7,8,21,22,37 while it dominates the unmutated
subset in CLL (Online Supplementary Figures S1A and
S1B).19Similarly, IGHV4-34 is used predominantly in a
mutated conformation by CLL, but was preferentially
unmutated in the HCL cases in our series (Figure 5A),
and even more significantly when all published HCL sequences were pooled (p=0.0002).2,7,8,21,22,37 Lack of
apparent HCDR3 stereotypy in HCL is another feature of variance with CLL, which, conversely, frequently groups cases with shared HCDR3 structural fea-tures.4,18,38 This also indicates that antigen drive may not
rely on HCDR3-mediated interactions in HCL. Con-versely, the low mutational rate of IGHV genes expressed in HCL, particularly of IGHV3-30 and
IGHV3-33 segments, which represent almost 50% of
all unmutated HCL, suggests that selective influences may be related to the IGHV segment itself.34,35
Analysis of Ig light chains provides novel evidence that HCL is characterized by selection events in the tumor BCR. In fact, HCL display: (i) an inverted Igκ:Igλ ratio (0.7:1); (ii) universal usage of IGLJ3 in the func-tional sIgλ expressors; and (iii) subsets with highly homologous KCDR3 and LCDR3. The preferential usage of Igλ light chain in our large series is consistent with prior independent studies,39-42and can be
consid-ered a unique feature of HCL. In the normal B-cell repertoire and in other B-cell neoplasms, Igκ is the most frequently used light chain.43-48 On these bases, our
results suggest that HCL requires selective usage of Igλ. The functional implications of Igλ selection in HCL remain speculative. The observations that (i) almost 50% HCL expressing sIgλ utilize IgHV3-21, IgHV3-30 or IgHV3-33 only; ii) virtually all HCL expressing Igλ utilize IgLJ3; and (iii) 40% HCL expressing Igλ display LCDR3 sets with shared structural features, suggest that HCL expressing Igλ may recognize common anti-gens requiring homologous LCDR3-J3 stretches.
LCDR3 identical motifs were documented within
IGHV3-21, IGHV3-30 or IGHV3-21 HCL (Table 1). In
public databases of normal or autoreactive B-cells, we
identified motifs identical to LCDR3 from HCL sets 1L or 2L, although specific antigen reactivities were found only in set L2.46The molecular triggers of the Igλ bias
and LCDR3-J3 selection in HCL are unknown. One possibility of IgL selection is that the Igκ-to-Igλ shift may derive from secondary rearrangements with res-cue of a new light chain in the periphery. Secondary rearrangements of Igλ after Igκ deletion have been observed in cases of Igλ-positive B-cell neoplasms.45-47,49
The observation of absent Ngc incorporations due to absent TdT activity in almost 50% HCL light chain rearrangements favors the hypothesis that secondary rearrangements occur in the periphery.27 We have
observed the potential ability of HCL to revise light chains in cases co-expressing mutated double light chain transcripts and/or protein with peripheral up-reg-ulation of RAG-1.50Also, double productive and
func-tional Ig light chain expression has been described in one independent case of HCL and is putatively the con-sequence of peripheral receptor revision.51
Classically, mutational status of IGHV genes distin-guishes whether the B-cell has encountered antigen in the germinal center with a T-cell-dependent reaction or whether it derives from antigen-naïve B cells.7
However, there is evidence that the BCR can also inter-act with antigen in a T-independent pathway and accu-mulate a low level of somatic mutations ectopically, likely in the marginal zones.21,52 Several observations
from both Ig heavy and light chain rearrangements indicate that HCL cells have experienced antigen stim-ulation. First, the majority of HCL are characterized by somatic mutations. Second, cloning of IGV region tran-scripts confirms the existence of low levels of intraclon-al heterogeneity intraclon-also in cases with 98% ≤ homology <100% to germline.19Third, the vast majority (82%) of
HCL co-express multiple pre-switch (IgM±D) and post-switch (IgG±A) sIgH, independently of mutational sta-tus and while activation-induced cytidine deaminase is expressed.2Histological findings and lack of CD27 and
CD38 suggest that, in typical HCL, the mutational and switch events are unlikely to occur in the germinal cen-Table 1. Intraclonal variability in hairy cell leukemia.
Code* IGHV Homology Intraclonal IGK/LV Homology Tumor/total IGL Intraclonal
(%) heterogeneity in (%) heterogenity in IGK/LV
in IGHV Unique Repeats HCL9 3-07 92.45 + KV1-D39(012/02) 97.49 11/11 3 0 HCL25/206 3-23 91.31 + KV3-11(L6) 97.13 6/11 1 0 HCL7/330 2-05 98.96 + KV3-20(A27) 96.90 9/12 3 0 HCL35 3-30 98.49 + LV1-44(1c) 97.16 4/12 4 1 HCL44/42 3-33 93.50 + LV1-47(1g) 94.68 7/7 2 0 HCL40 3-30 97.61 + LV1-47(1g) 98.93 11/12 2 0 HCL38/283 3-30 100.00 - LV2-11(2e) 100.00 10/10 0 0 HCL3/266 1-02 96.18 + LV2-14(2a2) 95.43 4/12 0 2 HCL68 4-39 93.84 + LV2-8(2c) 96.84 8/8 5 2 HCL27 3-23 96.52 + LV3-21(3h) 98.18 9/11 3 0 HCL32 3-30 95.83 + LV3-21(3h) 96.01 7/12 4 0 HCL26/216 3-23 91.66 + LV4-69(4b) 95.87 11/11 4 0
Intraclonal heterogeneity in the cloned IgHV products was defined positive when the same mutation was present in more than one clone from different isotype transcripts.2,7,29*The HCL cases with previously published details on intraclonal variation in tumor IgH isotypes are listed with the original code after the slash.2,7,29
ter.11 Additionally, in the present large series of HCL,
we observed an irrelevant incidence of novel N-glyco-sylation sites introduced by somatic mutation in the tumor IgV region (<6%), similar to that observed in normal memory B cells or in transformed memory and marginal zone B cells (Online Supplementary Tables S1
and S2).5,6,19,53 Since N-glycosylation sites are specifically
introduced in the IgV region of germinal center-derived lymphomas, while they are rare in marginal zone or post-germinal center B-cell neoplasms,5,6,19,53our results
provide further support to the hypothesis that HCL originates from non-germinal center derived B cells.
CD27-negative memory B cells or marginal zone B cells are the candidate counterparts of HCL.54
Comparison of the gene expression profiles of 14 HCL with the profiles of normal B cells from the naïve, ger-minal center, memory and plasma cell compartments showed that hairy cells were related to memory B cells.10However, the data were limited by the
unavail-ability of a marginal zone B-cell compartment.1,10
Indeed, HCL and marginal zone B cells share several immunogenetic features, including variable tiers of IGHV mutations, evidence of ongoing somatic hyper-mutation and lack of novel N-glycosylation sites.19,36,55
The morphology and phenotype of hairy cells also resemble those of marginal zone-derived CD23-nega-tive, CD27-negaCD23-nega-tive, CD38-negative interfollicular B cells. Overall, efforts should be directed at identifying
the normal counterpart within these categories.1,36,55
Knowledge of IGV gene use in these B-cell populations will be informative and perhaps more relevant when available from both normal and transformed B-cells.
In conclusion, the present largest immunogenetic sur-vey identifies the IG repertoire selected in HCL. The remarkable preference for Igλ with universal IGLJ3 use and a high incidence of LCDR3 homologous motifs fur-ther clarifies the selective influences present in the BCR of HCL. Whether this distribution of repertoire in HCL reflects that of a postulated normal B-cell counterpart remains to be clarified, and may follow from a more accurate characterization of normal B-cell subsets.
Authorship and Disclosures
FF designed the study, collected and processed the samples, performed experiments, analysed results and wrote the manuscript; ES, DR, SSS, TA, DR, FT and LL collected and processed the samples, performed exper-iments, and analyzed results; LT provided tumor sam-ples, performed experiments and analyzed data; GG designed the study, provided tumor samples, analyzed data and wrote the manuscript; FL provided tumor samples, supervised the project and manuscript writ-ing. The authors reported no potential conflicts of interest.
References
1. Tiacci E, Liso A, Piris M, Falini B. Evolving concepts in the pathogene-sis of hairy-cell leukaemia. Nat Rev Cancer 2006;6:437-48.
2. Forconi F, Sahota SS, Raspadori D, Ippoliti M, Babbage G, Lauria F, et al. Hairy cell leukemia: at the crossroad of somatic mutation and isotype switch. Blood 2004;104:3312-7. 3. Stevenson FK, Sahota SS,
Ottens-meier CH, Zhu D, Forconi F, Hamblin TJ. The occurrence and sig-nificance of V gene mutations in B cell-derived human malignancy. Adv Cancer Res 2001;83:81-16.
4. Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smi-levska T, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: patho-genetic implications and clinical cor-relations. Blood 2007; 109:259-70. 5. McCann KJ, Johnson PW, Stevenson
FK, Ottensmeier CH. Universal N-glycosylation sites introduced into the B-cell receptor of follicular lym-phoma by somatic mutation: a sec-ond tumorigenic event? Leukemia 2006;20:530-4.
6. Zhu D, McCarthy H, Ottensmeier CH, Johnson P, Hamblin TJ, Ste-venson FK. Acquisition of potential N-glycosylation sites in the immu-noglobulin variable region by somat-ic mutation is a distinctive feature of follicular lymphoma. Blood 2002;99: 2562-8.
7. Forconi F, Sahota SS, Raspadori D, Mockridge CI, Lauria F, Stevenson
FK. Tumor cells of hairy cell leu-kemia express multiple clonally related immunoglobulin isotypes via RNA splicing. Blood 2001;98:1174-81.
8. Maloum K, Magnac C, Azgui Z, Cau C, Charlotte F, Binet JL, et al. VH gene expression in hairy cell leu-kaemia. Br J Haematol 1998;101: 171-8.
9. Thorselius M, Walsh SH, Thunberg U, Hagberg H, Sundstrom C, Ro-senquist R. Heterogeneous somatic hypermutation status confounds the cell of origin in hairy cell leukemia. Leuk Res 2005;29:153-8.
10. Basso K, Liso A, Tiacci E, Benedetti R, Pulsoni A, Foa R, et al. Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhe-sion receptors. J Exp Med 2004;199: 59-68.
11. Forconi F, Raspadori D, Lenoci M, Lauria F. Absence of surface CD27 distinguishes hairy cell leukemia from other leukemic B-cell malignan-cies. Haematologica 2005;90:266-8. 12. Harris NL, Jaffe ES, Diebold J,
Flandrin G, Muller-Hermelink HK, Vardiman J, et al. The World Health Organization classification of neo-plastic diseases of the haematopoiet-ic and lymphoid tissues: eeport of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November 1997. Histopathology 2000;36:69-86.
13. Forconi F, King CA, Sahota SS, Kennaway CK, Russell NH, Ste-venson FK. Insight into the potential
for DNA idiotypic fusion vaccines designed for patients by analysing xenogeneic anti-idiotypic antibody responses. Immunology 2002;107: 39-45.
14. Giudicelli V, Chaume D, Lefranc MP. IMGT/V-QUEST, an integrated soft-ware program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res 2004;32:W435-440. 15. MRC ws. VBASE database. Available
at http://www.mrc-cpe.cam.ac.uk. 16. Matsuda F, Ishii K, Bourvagnet P,
Kuma K, Hayashida H, Miyata T, et al. The complete nucleotide se-quence of the human immuno-globulin heavy chain variable region locus. J Exp Med 1998;188:2151-62. 17. Kawasaki K, Minoshima S, Nakato
E, Shibuya K, Shintani A, Schmeits JL, et al. One-megabase sequence analysis of the human immunoglob-ulin l gene locus. Genome Res 1997; 7:250-61.
18. Messmer BT, Albesiano E, Efremov DG, Ghiotto F, Allen SL, Kolitz J, et al. Multiple distinct sets of stereo-typed antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med 2004;200:519-25.
19. Forconi F, Capello D, Berra E, Rossi D, Gloghini A, Cerri M, et al. Incidence of novel N-glycosylation sites in the B-cell receptor of lym-phomas associated with immunode-ficiency. Br J Haematol 2004;124: 604-9.
20. Brezinschek HP, Foster SJ, Brezin-schek RI, Dorner T, Domiati-Saad R, Lipsky PE. Analysis of the human
VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripher-al CD5(+)/IgM+ and CD5 (-)/IgM+ B cells. J Clin Invest 1997; 99: 2488-501.
21. Vanhentenrijk V, Tierens A, Wlo-darska I, Verhoef G, Wolf-Peeters CD. V(H) gene analysis of hairy cell leukemia reveals a homogeneous mutation status and suggests its marginal zone B-cell origin. Leukemia 2004;18:1729-32. 22. Arons E, Sunshine J, Suntum T,
Kreitman RJ. Somatic hypermuta-tion and VH gene usage in hairy cell leukaemia. Br J Haematol 2006;133: 504-12.
23. Kayano H, Dyer MJ, Zani VJ, Laffan MA, Matutes E, Asou N, et al. Aberrant rearrangements within the immunoglobulin heavy chain locus in hairy cell leukemia. Leuk Lymphoma 1994;14[Suppl 1]:41-7. 24. Foster SJ, Brezinschek HP,
Brezin-schek RI, Lipsky PE. Molecular mechanisms and selective influences that shape the k gene repertoire of IgM+ B cells. J Clin Invest 1997;99: 1614-27.
25. Farner NL, Dorner T, Lipsky PE. Molecular mechanisms and selec-tion influence the generaselec-tion of the human V lambda J lambda reper-toire. J Immunol 1999;162:2137-45. 26. Ignatovich O, Tomlinson IM, Jones
PT, Winter G. The creation of diver-sity in the human immunoglobulin V(lambda) repertoire. J Mol Biol 1997;268:69-77.
27. Nussenzweig MC, Shaw AC, Sinn E, Danner DB, Holmes KL, Morse HC 3rd, et al. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin mu. Science 1987;236:816-9. 28. Forconi F, Sozzi E, Raspadori D,
Toraldo F, Defina M, Zaja F, et al. Hairy cell leukemia (HCL) with unmutated V-genes have a poorer response to single agent 2CdA than HCL with mutated V-genes. Blood 2006;108:2327. Abstract 659a. 29. Forconi F, Sahota SS, Lauria F,
Stevenson FK. Revisiting the defini-tion of somatic mutadefini-tional status in B-cell tumors: does 98% homology mean that a V(H)-gene is unmutat-ed? Leukemia 2004;18:882-3. 30. Wilson PC, Wilson K, Liu YJ,
Banchereau J, Pascual V, Capra JD. Receptor revision of immunoglobu-lin heavy chain variable region genes in normal human B lympho-cytes. J Exp Med 2000;191:1881-94. 31. Kobrin C, Bendandi M, Kwak L.
Novel secondary Ig VH gene rearrangement and in-frame Ig heavy chain complementarity-determining region III insert-ion/deletion variants in de novo fol-licular lymphoma. J Immunol 2001; 166:2235-43.
32. Dorner T, Brezinschek HP, Bre-zinschek RI, Foster SJ, Domiati-Saad R, Lipsky PE. Analysis of the fre-quency and pattern of somatic mutations within nonproductively rearranged human variable heavy
chain genes. J Immunol 1997;158: 2779-89.
33. Crowley JJ, Mageed RA, Silverman GJ, Chen PP, Kozin F, Erger RA, et al. The incidence of a new human cross-reactive idiotype linked to subgroup VHIII heavy chains. Mol Immunol 1990;27:87-94.
34. Silverman GJ, Sasano M, Wormsley SB. Age-associated changes in bind-ing of human B lymphocytes to a VH3-restricted unconventional bac-terial antigen. J Immunol 1993;151: 5840-55.
35. Domiati-Saad R, Lipsky PE. Staphy-lococcal enterotoxin A induces sur-vival of VH3-expressing human B cells by binding to the VH region with low affinity. J Immunol 1998; 161:1257-66.
36. Lazzi S, Bellan C, Tiacci E, Palummo N, Vatti R, Oggioni M, et al. IRTA1+ monocytoid B cells in reactive lym-phadenitis show a unique topo-graphic distribution and immuno-phenotype and a peculiar usage and mutational pattern of IgVH genes. J Pathol 2006;209:56-66.
37. Thorselius M, Walsh SH, Thunberg U, Hagberg H, Sundstrom C, Ro-senquist R. VH gene analysis in hairy cell leukemia reveals preferen-tial VH3 family gene usage and het-erogeneous somatic hypermutation status. Blood 2003;102:893a [abstract].
38. Bomben R, Dal Bo M, Capello D, Benedetti D, Marconi D, Zucchetto A, et al. Comprehensive characteri-zation of IGHV3-21-expressing B-cell chronic lymphocytic leukemia: an Italian multicenter study. Blood 2007;109:2989-98.
39. Jansen J, Schuit HR, Hermans J, Hijmans W. Prognostic significance of immunologic phenotype in hairy cell leukemia. Blood 1984;63:1241-4. 40. Robbins BA, Ellison DJ, Spinosa JC, Carey CA, Lukes RJ, Poppema S, et al. Diagnostic application of two-color flow cytometry in 161 cases of hairy cell leukemia. Blood 1993;82: 1277-87.
41. Hansen DA, Robbins BA, Bylund DJ, Piro LD, Saven A, Ellison DJ. Identification of monoclonal noglobulins and quantitative immu-noglobulin abnormalities in hairy cell leukemia and chronic lympho-cytic leukemia. Am J Clin Pathol 1994; 102:580-5.
42. Arons E, Suntum T, Sunshine J, Stetler-Stevenson M, Kreitman RJ. Immunoglobulin light chain reper-toire in hairy cell leukemia. Leuk Res 2007;31:1231-6.
43. Langman RE, Cohn M. The propor-tion of B-cell subsets expressing kappa and lambda light chains changes following antigenic selec-tion. Immunol Today 1995;16:141-4.
44. Tumkaya T, van der Burg M, Garcia Sanz R, Gonzalez Diaz M, Langerak AW, San Miguel JF, et al. Immu-noglobulin l isotype gene rearrange-ments in B cell malignancies. Leukemia 2001;15:121-7.
45. Hadzidimitriou A, Stamatopoulos
K, Belessi C, Lalayianni C, Stavro-yianni N, Smilevska T, et al. Immunoglobulin genes in multiple myeloma: expressed and non-expressed repertoires, heavy and light chain pairings and somatic mutation patterns in a series of 101 cases. Haematologica 2006;91:781-7.
46. Stamatopoulos K, Belessi C, Hadzi-dimitriou A, Smilevska T, Kalagia-kou E, Hatzi K, et al. Immuno-globulin light chain repertoire in chronic lymphocytic leukemia. Blood 2005;106:3575-83.
47. Stamatopoulos K, Belessi C, Papadaki T, Kalagiakou E, Stavro-yianni N, Douka V, et al. Immuno-globulin heavy- and light-chain repertoire in splenic marginal zone lymphoma. Mol Med 2004;10:89-95.
48. Stamatopoulos K, Kosmas C, Papa-daki T, Pouliou E, Belessi C, Afen-daki S, et al. Follicular lymphoma immunoglobulin kappa light chains are affected by the antigen selection process, but to a lesser degree than their partner heavy chains. Br J Haematol 1997;96:132-46.
49. Perfetti V, Vignarelli MC, Palladini G, Navazza V, Giachino C, Merlini G. Insights into the regulation of immunoglobulin light chain gene rearrangements via analysis of the kappa light chain locus in lambda myeloma. Immunology 2004;112: 420-7.
50. Forconi F, Amato T, Raspadori D, Sahota SS, Dell’Aversano MA, Tassi M, et al. VH and VL genes in hairy cell leukemia reveal a dynamic ongoing modification of the surface B-cell receptor. Blood 2005;106:87a. 51. van der Burg M, Tumkaya T,
Boerma M, de Bruin-Versteeg S, Langerak AW, van Dongen JJ. Ordered recombination of immuno-globulin light chain genes occurs at the IGK locus but seems less strict at the IGL locus. Blood 2001;97:1001-8.
52. Lopes-Carvalho T, Kearney JF. Development and selection of mar-ginal zone B cells. Immunol Rev 2004;197:192-205.
53. Zhu D, Ottensmeier CH, Du MQ, McCarthy H, Stevenson FK. Inci-dence of potential glycosylation sites in immunoglobulin variable regions distinguishes between sub-sets of Burkitt’s lymphoma and mucosa-associated lymphoid tissue lymphoma. Br J Haematol 2003;120: 217-22.
54. Fecteau JF, Cote G, Neron S. A new memory CD27-IgG+ B cell popula-tion in peripheral blood expressing VH genes with low frequency of somatic mutation. J Immunol 2006; 177:3728-36.
55. Marafioti T, Jones M, Facchetti F, Diss TC, Du MQ, Isaacson PG, et al. Phenotype and genotype of interfol-licular large B cells, a subpopulation of lymphocytes often with dendritic morphology. Blood 2003;102:2868-76.