UNIVERSITÀ DEGLI STUDI DI ROMA
"TOR VERGATA"
FACOLTA' DI MEDICINA
DOTTORATO DI RICERCA IN
MICROBIOLOGIA MEDICA ED IMMUNOLOGIA
XXI CICLO
Multidrug resistance in Salmonella:
molecular characterization of a new emerging clone
of Salmonella enterica serotype Typhimurium
Dottoranda: Claudia Lucarelli
Tutor: Dr.ssa Ida Luzzi
Acknowledgments
I would like thank, my supervisor Dr. Ida Luzzi, Department of Infectious, Parasitic and Immuno-Mediated Diseases, and Dr. Alfredo Caprioli, Department of Veterinary Public Health and Food Safety, of Istituto Superiore di Sanita’ (ISS), Rome, Italy, which gave me the opportunity to do this PhD. course.
I would like thank too Dr. Laura Villa for her invaluable and fundamental scientific support during the course of the work and for critical review of this thesis as well. The consistent guidance and critical review has been a solid base for realization of this work. I would also like to thank many colleagues for their precious contributions to this work: in particular Anna Maria Dionisi, Ildo Benedetti, Emma Filetici, Slawomir Owczarek, Sergio Arena and Dr. Caterina Graziani. I also want to thank everyone of my laboratory for the lovely time spent all together and for their support in the practical difficulty, especially Dr. Alessandra Carattoli.
I would like thank Prof. John Threlfall and Dr. Katie Hopkins of Health Protection Agency, London, UK, and Dr. Mia Torpdhal of Statens Serum Institut, Copenhagen, Denmark, which provided some bacterial strains for this study, and for their helpful discussion concerning our future manuscript.
Finally I would like acknowledging my whole family, in particular the moral and practical assistance by my mother Maria Grazia, all my friends that support me during the course of this work, and Giovanni, for his costant loving backing.
Riassunto
Salmonella enterica sierotipo Typhimurium (STM) rappresenta la prevalente causa di gastroenterite trasmessa da alimenti in Italia, con la maggior parte degli isolati con resistenza multipla agli antibiotici, principalmente ad ampicillina (A), cloramfenicolo (C), streptomicina (S), sulfamidici (Su) e tetraciclina (T) (ACSSuT).
Un nuovo pattern di resistenza (R-type) ASSuT, mancante della resistenza a C, è recentemente emerso in Italia tra ceppi di STM e della sua variante monofasica, Salmonella enterica subspecie enterica sierotipo S. 4,[5],12:i:– .
L’obiettivo principale di questa tesi è stato la caratterizzazione di ceppi di STM e di S. 4,[5],12:i:– con R-type ASSuT, usando tecniche di tipizzazione molecolari e fenotipiche, quali l’elettroforesi in campo pulsato (PFGE) e la fagotipizzazione, allo scopo di valutare la loro origine clonale e la loro relazione con i ceppi ACSSuT. Usando il database Pulse-Net Europe è stata valutata la presenza di ceppi ASSuT in altre nazioni Europee al fine di allestire una collezione internazionale di ceppi. Questa collezione è stata ulteriormente caratterizzata, identificando i geni di resistenza, investigando la loro localizzazione, e determinando la regione di resistenza.
Sia tra ceppi di STM che di S. 4,[5],12:i:–, il principale profilo di PFGE è rappresentato da STYMXB.0079, mentre i ceppi STM ACSSuT appartengono ai profili STYMXB.0061 e STYMXB.0067. L’analisi dei profili di PFGE con il software Bionumerics ha mostrato che più del 90% dei ceppi ASSuT e ACSSuT appartenevano a due distinti clusters con un’omologia genetica del 73%, dati che dimostrano l’appartenenza dei ceppi ASSuT ad un'unica linea clonale differente da quella dei ceppi ACSSuT. La maggior parte dei ceppi con profilo ASSuT non erano tipizzabili (DTNT) attraverso la fagotipizzazione o appartenevano al fagotipo U302. Al contrario i ceppi ACSSuT appartenevano principalmente al fagotipo DT104.
Successivamente, nel database Pulse-Net Europe, è stato possibile identificare ceppi ASSuT, sia STM che S. 4,[5],12:i:– isolati in Danimarca ed Inghilterra, con profili di PFGE identici o strettamente correlati a quelli dei ceppi italiani, dati che indicano che il clone ASSuT è presente anche in altri paesi Europei. Al fine di identificare i geni responsabili della resistenza sono stati selezionati 64 ceppi di STM e S. 4,[5],12:i:– ASSuT, e 11 ceppi di STM con differenti R-type e profili di PFGE, usati come controlli. Tutti i ceppi provenivano da infezioni umane ed erano stati isolati in Italia, Danimarca e Inghilterra.
Tutti i ceppi ASSuT erano positivi per i seguenti geni di resistenza: blaTEM, strA-strB, sul2 and tet(B).
Successivi esperimenti di localizzazione hanno dimostrato che i geni di resistenza ASSuT sono localizzati sul cromosoma
Infine, è stata determinata la sequenza completa del cluster di resistenza ASSuT. Questo cluster è composto da due isole di resistenza (RI1 e RI2) separate da DNA cromosomale. In particolare, RI1 è compresa tra due IS26 e contiene ∆tnp3R, blaTEM-1, tnpB, seguito da strB, strA, sul2, repC, ∆repA ed
un’atra IS26. RI2 è anch’essa compresa tra due IS26, comprendenti ∆IS10L, il gene di resistenza alla tetraciclina, IS1, l’operone per la resistenza al mercurio, il gene yaeA, e un’ ipotetica transposasi (tniA∆). Entrambe le RIs mostrano il 99% di identità con due regioni adiacenti del plasmide pHCM1, presente nel ceppo di S. Typhi isolato in Vietnam. Le sequenze di inserzione IS26 potrebbero aver avuto un ruolo nella formazione di questo cluster di resistenza, ma quest’ipotesi deve essere ancora verificata.
In conclusione il lavoro di questa tesi indica che i ceppi ASSuT di STM e S. 4,[5],12:i:–, in aumento in Italia, appartengono ad un'unica linea clonale e che i ceppi S. 4,[5],12:i:– circolanti nella nostra nazione, derivano principalmente da questa linea clonale di STM. Inoltre il clone ASSuT è diffuso anche in Danimarca ed Inghilterra. Il pattern di antibiotico resistenza conferito da un’isola di resistenza cromosomale, con un’organizzazione simile ad altri cluster precedentemente descritti, suscita preoccupazione poichè la resistenza può essere mantenuta stabilmente in assenza di pressione selettiva.
Keywords
Salmonella Typhimurium, Salmonella 4,[5],12:i:–, Multidrug Resistance, Pulsed Field Gel Electrophoresis, Phage Typing, Chromosomal Resistance Island.
Abstract
Salmonella enterica serovar Typhimurium (STM) represents the prevalent cause of foodborne gastroenteritis in Italy with the majority of isolates exhibiting multidrug resistance, mainly to ampicillin (A), chloramphenicol (C), streptomycin (S), sulfonamide (Su) and tetracycline (T) (ACSSuT).
However, a new resistance pattern (R-type) ASSuT, lacking resistance to C, has recently emerged in Italy among strains of STM and of its monophasic variant, Salmonella enterica subspecie enterica serovar S. 4,[5],12:i:– .
The main objective of this thesis has been the characterization of STM and S. 4,[5],12:i:– strains with R-type ASSuT, using both molecular and phenotypic typing technique, pulsed-field gel electrophoresis (PFGE) and phage typing, in order to evaluate their clonal origin and the relationships with the ACSSuT strains. In addition, by the use of the Pulse-Net database it was evaluated if ASSuT strains were present in other European countries in order to set up an international collection of these strains. This collection has been further characterized with the identification of resistance genes, the investigation of their localization, and determination of the resistance region.
Among both the STM and S. 4,[5],12:i:– ASSuT strains, the predominant PFGE profile was STYMXB.0079, while the STM ACSSuT strains belonged to the STYMXB.0061 and STYMXB. 0067. Bionumerics cluster analysis of PFGE profiles showed that more than 90% of ASSuT and ACSSuT resistant strains were included in two distinct clusters with a genetic homology of 73% each other, suggesting that the ASSuT resistant strains belong to a same clonal lineage different from that of the ACSSuT strains.
Phage typing showed that both STM and S. 4,[5],12:i:– ASSuT strains were not typeable (DTNT) or U302. A different figure was observed for the ACSSuT strains: the STM strains mostly belonged to DT104.
The Pulse-Net Europe database, allowed us to identify ASSuT strains, both STM and S. enterica 4,[5],12:i:–, isolated in Denmark and UK, with the same or very closely related PFGE patterns as the Italian strains, suggesting that the ASSuT clone is circulating in different European countries.
The resistance genes were identified in 64 strains of STM and S. enterica 4,[5],12:i:–with ASSuT R-type and in 11 STM strains with different resistance patterns and PFGE profiles as controls. All strains were isolated from human infections in Italy, Denmark and UK.
All the ASSuT strains were positive for the following resistance genes: blaTEM, strA-strB, sul2 and tet(B).
The control strains showed the same gene pattern, in accordance with their resistance profiles. A variability of the genes conferring resistance to tetracycline was detected. Localization experiments demonstrated that the ASSuT resistance genes are chromosomally located.
Finally, the complete sequence of ASSuT resistance cluster was determined. This cluster is composed by two resistance island (RI1 and RI2) divided by chromosomal DNA. In particular, RI1 is comprised between two IS26 and contains ∆tnp3R, blaTEM-1, tnpB , followed by strB, strA, sul2, repC, ∆repA and
another IS26. RI2 is bracketed by two IS26, comprising ∆IS10L, tetracycline resistance gene, IS1, the operon for resistance to mercury, yaeA gene, and a putative transposase (tniA∆). Both this RIs show 99% sequence identity to two adiacent region of pHCM1 plasmid, harbored in S. Typhi isolated in Vietnam. IS26 elements could have played a role in the assembly of this resistance cluster but it will be investigated more in detail.
In conclusion the work of this thesis indicates that the tetra-resistant ASSuT strains of STM and S. 4,[5],12:i:–, increasingly isolated in Italy, belong to a same clonal lineage and that the S. 4,[5],12:i:– strains circulating in our country, mainly derive from this STM clonal lineage. ASSuT clone is also circulating in Denmark and United Kingdom. The antimicrobial resistance pattern conferred by a chromosomal island, with an organization similar to previously reported clusters, deserves concern since
Index
1
.Salmonella spp.
1
2. Infections of Salmonella in humans
4
3. Epidemiological features
6
4. Surveillance of salmonellosis
8
4.1 The Enter-Net International Surveillance Network 9
4.2 Enter-Net Italia 10
5. Epidemiological Typing methods
11
5.1 Phage typing 11
5.2 Molecular typing methods 11
5.2.1 Pulsed field gel electrophoresis (PFGE) 13
6. Antimicrobial resistance in Salmonella
15
7.
Objective
18
8. Materials and Methods
19
8.1 Bacterial strains 19
8.2 Serotyping 19
8.3 Antimicrobial susceptibility test 19
8.4 Phage typing 20
8.6 Identification of resistance genes 22
8.6.1 Total genomic DNA purification 22
8.6.2 Polymerase Chain Reaction assays (PCR) 23
8.6.3 DNA sequencing 23
8.7 Plasmidic and chromosomal investigation 24
8.7.1 Plasmid DNA purification 24
8.7.2 Transfer experiments 25
8.7.3 PFGE with I-Ceu I 25
8.7.4 Southern blot hybridization experiments 25
8.8 Cosmid library 26
9. Results
28
9.1 Antimicrobial resistance pattern in Salmonella Typhimurium
and its monophasic variant 28
9.2 PFGE profiles of ASSuT strains 29
9.3 Phage typing of ASSuT strains 32
9.4 Identification and localization of resistance genes 35 9.5 Genetic characterization of chromosomal resistance island ASSuT 41
10. Discussion
45
1. Salmonella spp.
The genus Salmonella, member of Enterobacteriaceae family, is formed up bacteria gram-negative, facultative anaerobic and peritrichously flagellated rods.
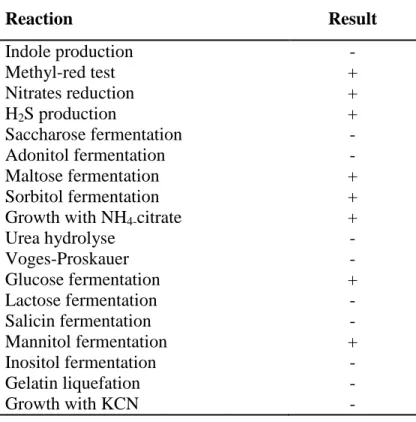
This genus is distinguished from members of other genera of Enterobacteriaceae by a series of biochemical reactions (Table 1) including the production of hydrogen sulfide, the reduction of nitrates, the decarboxylation of lysine and ornithine and the utilization of citrate (Andrews and Baumler, 2005).
Table 1: Biochemical reactions of Salmonella genus
Reaction Result Indole production - Methyl-red test + Nitrates reduction + H2S production + Saccharose fermentation - Adonitol fermentation - Maltose fermentation + Sorbitol fermentation +
Growth with NH4-citrate +
Urea hydrolyse - Voges-Proskauer - Glucose fermentation + Lactose fermentation - Salicin fermentation - Mannitol fermentation + Inositol fermentation - Gelatin liquefation - Growth with KCN -
The Salmonella genus includes two species: Salmonella bongori and
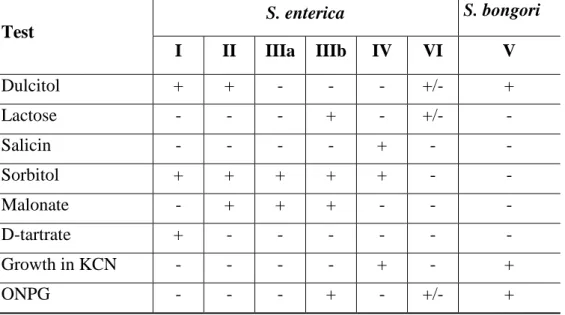
On the basis of biochemical assay (Table 2), Salmonella enterica spp. can be distinguished into six subspecies:
I. S. enterica subspecie enterica
II. S. enterica subspecie salamae
IIIa. S. enterica subspecie arizonae
IIIb. S. enterica subspecie diarizoane
IV. S. enterica subspecie houtenae
V. S. enterica subspecie indica
Table 2: Biochemical reactions for differentiating the seven Salmonella subspecies Test S. enterica S. bongori I II IIIa IIIb IV VI V Dulcitol + + - - - +/- + Lactose - - - + - +/- - Salicin - - - - + - - Sorbitol + + + + + - - Malonate - + + + - - - D-tartrate + - - - - Growth in KCN - - - - + - + ONPG - - - + - +/- +
Salmonella enterica subspecie enterica has particular importance in human
and animal infections and its classification has been further developed by using technique of serologic typing, based on the presence or absence of somatic (O), capsular (Vi), and flagellar (H) antigens (Yan et al., 2003).
The O antigen is a heat-stable polysaccharides that have a core structure, lipopolysaccharide (LPS), that is common to all enterobacteria, and side chains of sugars attached to the core determine O specifity. Sixty-five different O antigens
are known, which are indicated by Arabic numerals (Holmes, 1998, Old et al., 1998).
The Vi antigen is a capsular polysaccharide that confers increased virulence and may be present in only three serotypes: S. Typhi, S. Paratyphi C and
S. Dublin (Old et al., 1998).
The flagellar antigens (H), indicated with small letter, are present only in mobile species of Salmonella. In most salmonellae the flagellar antigens exist in 2 alternative phases: phase 1 (or specific phase, typical of one serotype) and phase 2 (or aspecific, common to more serotypes), that represent two distinct protein encoded by 2 genes, respectively fliC and fljB (Yan et al., 2003). The variation of phases is determined by an invertable element (hin) containing the promoter region of the phase operon. With this invertible sequence in one orientation, the promoter is correctly positioned to allow reading of the fljB gene (phase 2) along with the repressor, fljA, that switches off the phase-1 gene. In the other orientation, the phase-2 gene cannot be read, the repressor of phase-1 gene is not formed and the flagella with phase-1 antigenicity are formed. Salmonella switches phases at characteristic frequencies and the transition can be induced in the laboratory by their cultivation in semi-solid agar containing antiserum against phase-1 or phase-2 flagellar antigens (Therfall, 2005; Echeita et al., 2002).
With the identification of these three antigens it is possible to distinguish above 2500 serotypes inside Salmonella genus. Serotypes are designated with the antigenic formula or with the “name” Salmonella, followed by the serotype (Le Minor, 1992).
Among the serotypes we can distinguish typhoidal Salmonella and non-typhoidal Salmonella (NTS). Only three serotypes, S. Typhi, S. Paratyphi C and S. Dublin, belongs to the first group. These serotypes are transmitted from person to person through oral-fecal cycle and are cause of an acute systemic infection named as typhoid fever.
All NTS are etiological agent of zoonoses, diseases and/or infections that can naturally transmit between vertebrate animals and man.
2. Infections of Salmonella in humans
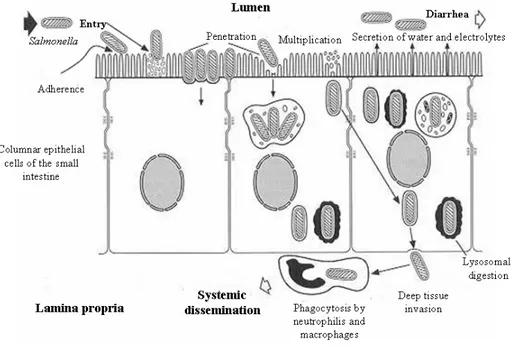
The pathogenetic process of salmonellosis (Figure 1) begins immediately after ingestion, when the organisms reach the intestinal lumen, replicate, adhere to enterocytes and invade the mucosa. Subsequently salmonellae become incorporated into phagesomes and without any alteration reached the lamina propria where they replicate quickly, causing inflammation with congestion, edema and influx of polymorphonuclear and monocytes-macrophages.
In this first stage, the release of bacterial lipopolysaccharide (LPS) causes fever, nausea, vomiting and abdominal pain. Diarrhea starts approximately 10-20 hours after: it is caused both by degeneration and detachment of enterocytes and their functional suffering. This biphasic evolution of symptoms occurs in most patients with Salmonella gastroenteritis. Faeces are initially soft and watery, and in the second or third day of illness traces of mucus mixed with blood can be present in stool (Tauxe et al., 1998, Darwin and Miller, 1999).
Symptoms usually regress in 2-4 days and in most cases the healing is complete, but the subject could remain carrier and excret Salmonella in the faeces, for over a month following infection.
Among the individuals with gastrointestinal illness, approximately 5% develops invasive Salmonella infections. The evolution of illness depends on both virulence of the bacteria and immune system of the host. The defensive capabilities are not very effective in infants, elderly and immunocompromised patients (Darwin and Miller, 1999; Yan et al., 2003).
Salmonella virulence depends on different factors, necessary to implement
all the various stages of infection (Wallis et al., 2000): survival in the gastric acid (Slauch et al., 1997), colonization of the intestine, adhesion to the cells of the intestinal lumen (fimbriae type 1 and 3) (Bäumler et al., 1997), invasion of the intestinal epithelium at the level of Peyer’s patches and survival in macrophages
An important role in the virulence is played by exogenous gene clusters located in the chromosome as SPI (Salmonella Pathogenicity Island), the hil gene (hyperinvasion locus), and virulence plasmids (Darwin and Miller, 1999, Lucas et al ., 2000).
Figure 1: Pathogenetic mechanism in Salmonella infection.
(modified from
http://www.surrey.ac.uk/SBMS/ACADEMICS_homepage/mcfadden_johnjoe/img /salmonella-epithelial%20interactions.jpg).
3. Epidemiological features
Salmonellosis are the 2nd most commonly reported human zoonoses in Europe: in 2006 a total of 168639 confirmed cases of human salmonellosis has been reported by TESSy, The European Surveillance System (http://ecdc.europa.eu/en/files/pdf/Publications/081215_AER_long_2008.pdf).
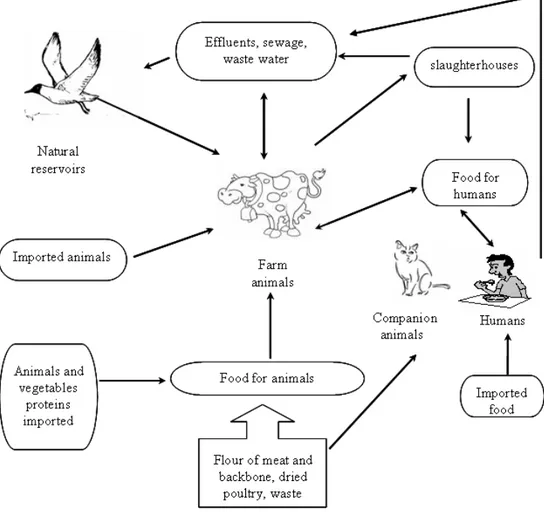
The epidemiological cycle of Salmonella (Figure 2) is of remarkable complexity involving animals reservoir, food and environment (van den Bogaard et al., 2000).
The animal reservoirs are mainly represented by food animals, such as swine, poultry and cattle. A considerable proportion of infected animals are asymptomatic carriers and the carriage of Salmonella by apparently healthy farm animals is directly responsible for their subsequent introduction into the derived food products (Andrews and Baumler, 2005).
The contaminated food products represent the most important vehicle by which people become infected with NTS. Food of vegetable origin and environment, that can be contaminated from faeces of human and animal origin, are important in the epidemiological Salmonella cycle, as well waste water, which are responsible for the spread of Salmonella in aquatic environments such as streams, rivers, lakes and could be a source of contamination of soil and plants (Lemarchand et al., 2002).
4. Surveillance of Salmonellosis
Salmonella infections are still representing an important public health issue in industrialized countries. Increasing international travel and current manufacturing and distribution practices play an important role in the occurrence of foodborne infections. All parts of the globe are now accessible within 24 hours, a period of time lesser than the incubation of most enteric diseases. Foodstuffs, manufactured or harvested at one place are distributed within a country or across economic regions (such as the EU), continents, or even worldwide. Events in one country, which previously might not have had implications outside its borders, now have the potential to affect many other countries. Outbreaks of infection may occur far from the source of contamination: free movement of people and goods between countries are effective ways of distributing disease internationally.
In Italy Salmonella infections are subject of mandatory notification (class II of the infectious diseases) to National Health Service (NHS), which registers about 10000 cases of salmonellosis each year. The main advantage of this statutory system is the stability during the time: however in many cases detailed microbiological information are lacking. Informations on Salmonella serotypes are of fundamental importance to understand, for istance, if an outbreak has been caused by identical or different strains.
Various surveillance systems, both at national and international level, formed by microbiological laboratories, involved in the identification and typing of strains, and epidemiologist, has been set up in order to early identify diseases and their causes (Fisher, 1999).
4.1 The Enter-Net International Surveillance Network
An international laboratory-based surveillance network for human gastrointestinal infection known with the acronym Enter-Net, was established by the European Commission (EC). To Enter-Net participated all European countries plus 6 non EU countries. Enter-Net activities, has been coordinated until 2007 by Health Protection Agency, London, United Kingdom, and since October 2007 it has been subsumed into The European Center for Disease Prevention and Control (ECDC), in the framework of Foodborne and Waterborne Zoonotic Diseases Network.This network monitors the incidence of salmonellosis, Verocytotoxin-producing Escherichia coli (VTEC), campylobacteriosis, listeriosis and other enteric pathogens.
The surveillance includes harmonization of the work of national reference laboratories, extension of the activities where necessary, and the pooling of the resultant data in a timely way, to create an international database. All the data collected are analyzed in order to identify and investigate outbreaks (Fisher, 1999).
As far as Salmonella is concerned, the network involves, other than collection of the isolates, their further characterization with serotyping, antimicrobial resistance tests and molecular technique by standardized protocols.
In addition, the network facilitates the study of resistance mechanisms and their genetic control by arranging a collection of representative strains of multi drug resistance (MDR) salmonellas and co-ordinating the required research work between specialised centres, and, where available, by comparing the resistances of isolates of different origin (Fisher, 1999).
4.2 Enter-Net Italia
Italy participates to the international Foodborne and Waterborne Disease Network through a national surveillance system, named Enter-Net Italia. To the network, coordinated by the Istituto Superiore di Sanità, collaborated about 100 peripherical Diagnostic Laboratories and 37 Regional Reference Laboratories (Galetta et al., 2006). Enter-Net Italia collects salmonella isolates from human cases and environmental sources and data are available at the website
http://www.iss.it/ente/.
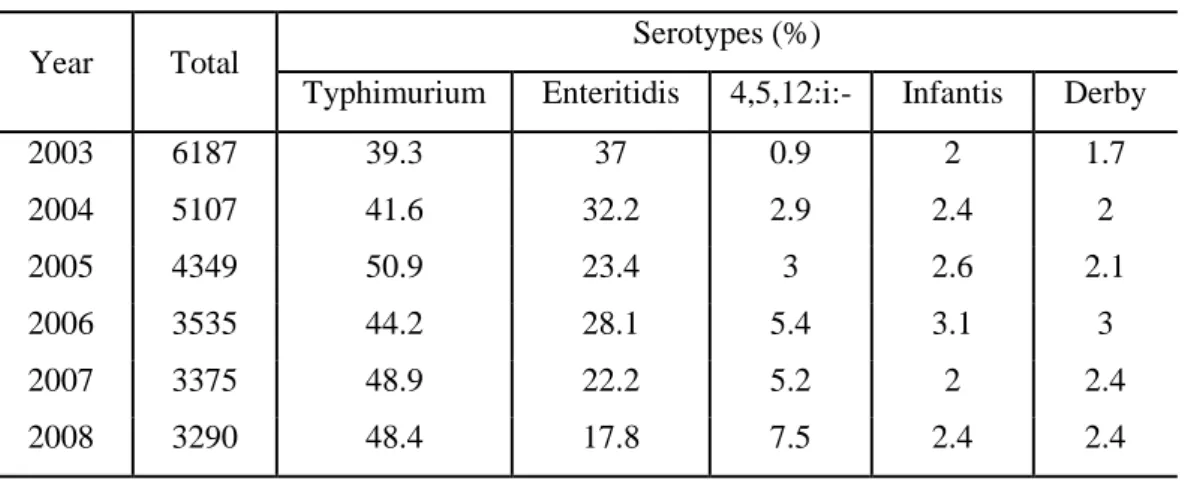
Aim of the surveillance system is to monitor the trend of Salmonella serotypes and the analysis of data indicates that over the last five years (Table 3), the main serotype isolated is represented by S. Typhimurium, followed by S. Enteritidis. In addition, it is possible to highlight that while in 2003 the frequency of isolation of S. 4,[5] ,12:i:– accounted for 0.9%, in 2008 its frequency reached 7.5%. An increase of this serotype has also been recorded in other European countries as Spain, Germany and England and Wales (Meakins et al., 2008).
Table 3: Top five serotypes among non-typhoidal salmonella strains isolated from
humans in Italy, during the years 2003-2008.
Year Total Serotypes (%)
Typhimurium Enteritidis 4,5,12:i:- Infantis Derby
2003 6187 39.3 37 0.9 2 1.7 2004 5107 41.6 32.2 2.9 2.4 2 2005 4349 50.9 23.4 3 2.6 2.1 2006 3535 44.2 28.1 5.4 3.1 3 2007 3375 48.9 22.2 5.2 2 2.4 2008 3290 48.4 17.8 7.5 2.4 2.4
5. Epidemiological Typing Methods
5.1 Phage Typing
Phage typing is a phenotypic technique that allows to distinguish strains belonging to the same serotype, and it is based on susceptibility of salmonellae towards a set of bacteriophages (Anderson et al., 1977). More in detail, the phage typing is based on the feature that some strains have receptors for some lytic phages, which can enter and replicate in the cell, inducing lysis (Yan et al., 2003). The phage typing represent a very useful technique when applied to the most common serotypes, S. Typhimurium and S. Enteritidis. Nevertheless the technique is too much depending on the interpretative ability of the operator, and it is difficult to standardize. It also required the maintenance of biologically active phage stocks, which are provided exclusively by Health Protection Agency (HPA, Colindale, London, UK) to National Reference Center (Yan et al., 2003).
Finally, the phage type is an instable character: in fact a determinate phage type could be converted in another (phage conversion) (Old, 1990). In S. Typhimurium this phenomenon seems to be caused by chromosomal mutation, by the acquisition or loss of resistance plasmid and by the integration of lysogen phages in the bacterial chromosome (Threlfall et al., 1978; Mmolawa et al., 2002).
5.2 Molecular Typing Methods
To supplement the more traditional phenotypic typing methods, a range of molecular typing techniques, based on characterization of the genotype of the organism by analysis of plasmid and chromosomal DNA, have been developed.
The use of these techniques is of primary importance in Salmonella to further differentiate strains belonging to serotypes for which phage typing is not available, and among strains showing the same phage type.
The molecular techniques, when compared with traditional methods, have the advantage of rapidity, allowing a rapid detection of outbreaks, and reliability, because of the direct analysis of the DNA (Peters et al., 2003).
Among molecular typing methods, five chromosomally based methods have been used for Salmonella: ribotyping; random cloned chromosomal sequence (RCCS) typing; insertion sequence (IS) 200 typing; pulsed-field gel electrophoresis (PFGE); and PCR-based methods such as random amplified polymorphic DNA typing (RAPD), amplified fragment length polymorphisms (AFLP) and variable number of tandem repeats fingerprinting (VNTR) (Threlfall, 2005).
The first three methods use DNA-DNA hybridization to identify restriction fragment length polymorphisms (RFLP) following detection of single or multicopy gene sequences across the bacterial genome; the fourth method (PFGE) is a modification of conventional agarose gel electrophoresis that allow analysis of the whole bacterial genome on a single gel.
The PCR-based methods are based on amplification of specific DNA sequence by PCR to produce characteristic group of fragments dependent upon the genome content of the strain.
The methods used most extensively in support of epidemiological investigations of Salmonella are: rybotyping, RCCS typing, IS200 fingerprinting, PFGE and VNTR. Currently, among these methods, PFGE is considered as the “gold standard” for subtyping of Salmonella isolates (Peters et al., 2003).
5.2.1 Pulsed-field gel electrophoresis
This technique, made up in 1983 by Schwartz and Cantor (1984), uses rare cutting restriction enzymes, which produce restriction fragments of a size greater than 30 kb. These fragments are separated under conditions with alternating electric field. This approach can resolve DNA fragments up to 800 kb in size.
By PFGE a restriction profile is obtained which identifies, in an univocal way the strains. The comparison of profiles obtained from different strains, permits to establish their genetic relationship (Gautom, 1997). The determination of genetic relatedness based on PFGE banding patterns had been standardized by Tenover et al., (1995): strains are designated genetically indistinguishable if their restriction patterns have the same numbers of bands and the corresponding bands are the same apparent size. Isolates which differs for two-three bands are defined closely related, for 4-6 bands are possibly related and isolates different for at least 7 bands are considered unrelated.
PFGE is highly reproducible and strongly discriminatory and has been shown to be highly effective in epidemiological studies conducted on different serovars of S. enterica (Harbottle et al., 2006; Herrero et al., 2006; Michael et al., 2006).
PFGE represents the basis of a molecular surveillance network for food-borne infections in Europe, the Pulse-Net Europe, which has the aim to establish a real-time linked surveillance database system to detect infection clusters and investigate outbreaks of Salmonella, and other important enteric pathogens, as verocytotoxigenic E. coli (VTEC) and Listeria monocytogenes
(http://www.pulsenet-europe.org/).
This network aims to establishes a standardized protocol of PFGE and a web-based central Pulse-Net Europe database, that makes possible for partners to submit the generated and analysed PFGE data directly via the web. In addition all the partners have direct access to the database and can compare their PFGE data
to the data that are stored in the database. The direct access ensures a uniform naming of PFGE-subtypes, on behalf of all participating countries, provides portable data and makes easy for partners to check their own data when alerts of infections clusters are posted on the Pulse-Net Europe communication forum (http://www.pulsenet-europe.org/).
The direct access to comparable typing data for isolates from different origin improves the surveillance and help tracing-back in food-borne infections at national, European and international level. Furthermore, international clusters of food-borne infections that have too few cases in each country to be detected by the National surveillance systems, could be detected through central Pulse-Net Europe surveillance systems. The data collected are also comparable internationally through Pulse-Net International (Pulse-Net Asia-Pacific region, Pulse-Net Canada, Pulse-Net Latin America and Pulse-Net USA).
6. Antimicrobial resistance in Salmonella
In human salmonellosis, antibiotic therapy is not indicated in case of gastroenteritis, except for people at risk of developing systemic disease such as children, elderly and immune compromised patients (Allen and Poppe, 2002). The usage of antibiotic in human therapy and the intensively usage of antibiotics in veterinary medicine, could have contributed to an increase in the occurrence of antimicrobial resistance among Salmonella isolates of several countries in recent years.
In Europe, in 2000, 35.2% of all non-typhoidal isolates were resistant to at least one antimicrobial; by 2005 this proportion had risen to 40.8%. Of particular concern is the significant increase in the number of isolates with resistance to 4 or more drugs (MDR), from 10.9% in 2000 to 15.2% in 2005 (Threlfall et al., 2003; Enternet annual report, 2005). In particular the resistance has been reported towards antimicrobial as ampicillin, sulfonamides tetracyclines and streptomycin, antimicrobial used from a long time in human and veterinary therapy (Threlfall et al., 2003).
Because of the increased resistance to conventional antibiotics, extended-spectrum cephalosporins and fluoroquinolones have become the drugs of choice for the treatment of infections caused by multidrug-resistant Salmonella serotypes, but resistances to these drugs are now a matter of concern (Threlfall et al., 2003; Allen and Poppe, 2002; Dionisi et al., 2009).
Moreover, human infections caused by antimicrobial resistant Salmonella have an higher fatality rate, and are more likely associated to hospitalization, usually for a longer time period than patients with infection caused by susceptible strains (Emborg et al., 2008).
The antimicrobial resistance rates are variable for the different antibiotics and for the different serotypes. S. Enteritidis, one of the most prevalent serotypes, is relatively more susceptible to antimicrobial agents than other serotypes. A high
rate of resistance is found in S. Typhimurium, another globally prevalent ubiquitous serotype. In United Kingdom, during the 80s, a multidrug-resistant strain of STM belonging to phage type 104 (DT104) was isolated and found to be simultaneously resistant to 5 antimicrobial agents, ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline (R-type ACSSuT). Subsequently this clone became widespread over the world (Su et al., 2004; Threlfall et al., 2002).
Molecular typing by PFGE has demonstrated that the majority of ACSSuT strains reported over the world belong only to two or three PFGE profile closely related, thus demonstrating the clonal origin of the strains (Baggesen et al., 2000; Threlfall, 2000; Threlfall et al., 2002).
Genetic studies have demonstrated that the ACSSuT resistance profile of
S. Typhimurium DT104 is chromosomally encoded: so resistance can be stably
maintained even in the absence of selective pressure (Cloeckaert and Schwarz., 2001).
The ACSSuT gene cluster is localized to a 13-kb segment of the
Salmonella Genomic Island I (SGI1), a 43-kb genomic island, which has 44 open
reading frames (ORFs). The antibiotic resistance region is composed by two class 1 integron, carrying a single resistance gene cassette in addition to the sul1 and
qacE
∆
1 genes, characteristics of this class 1 integron, and specifying resistance tosulfonamides and disinfectant respectively. The first integron carrying aadA2 gene confers resistance to streptomycin and the second integron containing the β-lactamase gene blaPSE-1 encodes for resistance to ampicillin. Between the two
integrons, the genes floR and tet(G) are located, conferring resistance respectively to chloramphenicol and tetracycline (Boyd et al., 2000).
Several hypothesis has been done on the origin of the DT104 antibiotic resistance gene cluster and consecutive spread of multidrug-resistant strains. The use of antimicrobial agents in agriculture might have contributed to the emergence of resistant DT104 strains. Since the genes included in the multidrug-resistance gene cluster of DT104 strains confer multidrug-resistance to four of the drugs
most frequently used in veterinary medicine (tetracyclines, β-lactams, aminoglycosides and sulfonamides), co-selection of the entire cluster may have been resulted from the use of any of these drugs. While some genes in the cluster, such as aadA2, blaPSE-1, or sul1, are widely distributed among
Enterobacteriaceae, the remaining two genes, floR and tet(G), are most unlikely
of enterobacterial origin. On the contrary they seem to derive from two bacterial species pathogenic for fishes: Photobacterium damselae and Vibrio anguillarum (Zhao and Aoki, 1992, Kim and Aoki, 1996). Therefore, it has been assumed that these resistance determinants have originated in aquatic bacteria and then horizontally transferred to S. Typhimurium DT104 (Angulo et al., 2000). To confirm this hypothesis the mobility of SGI1 has been demonstrated and this ability could have contributed to the spread of this genomic island among different S. enterica serotypes (Mulvey et al., 2006). It is also important to note that MDR DT104 strains seem to be more virulent, despite specific component participating in virulence remain to be identified.
Although ACSSuT clone is diffused also in Italy, since 2000, strains of STM with a tetra-resistant pattern ASSuT, with or without additional resistances, but lacking resistance to chloramphenicol, have emerged either among human and food animals isolates (Busani et al., 2004). In particular, while the frequency of the ACSSuT pattern among the Italian human isolates of STM remained constant over the years accounting for 29.6 % in 2006, the frequency of the tetra-resistant pattern increased from 7% in the period 1999-2001 to 34.1% in 2006 (Busani et al., 2004; Graziani et al., 2008).
In Italy the tetra-resistant pattern is also prevalent (Barone, et al., 2008; Galetta et al., 2008) among human isolates belonging to Salmonella enterica subspecies serovar 4,[5],12:i:– (S. 4,[5],12:i:– ), defined as a monophasic variant of S. Typhimurium. This serovar shares almost all antigenic factors with STM, but it lacks the second-phase flagellar antigen encoded by the fljB gene (Zamperini et al., 2007; Echeita et al., 2001).
7. Objective
The main objective of this thesis has been the characterization of both STM and S. 4,[5],12:i:– strains with R-type ASSuT.
ASSuT strains were characterized using both molecular and phenotypic typing technique, pulsed-field gel electrophoresis and phage typing respectively, in order to evaluate their clonal origin and the relationships with the ACSSuT strains.
In addition, by the use of the Pulse-Net database, it was evaluated if ASSuT strains were present in other European countries in order to set up an international collection of these strains. This collection has been further characterized by the identification of resistance genes, the investigation of their localization, and determination of the resistance region.
8. Materials and methods
8.1 Bacterial strains
STM and S. 4,[5],12:i:– strains were selected for the different phases of the study from the collection of the National Reference laboratory at the Istituto Superiore di Sanità, Rome, for the Enter-Net Italia surveillance.
Danish and English strains included in this study have been provided by Statens Serum Institute, Copenhagen, Denmark, and by Health Protection Agency, London, United Kingdom.
8.2 Serotyping
Serotyping of Salmonella spp. isolates was performed for the identification of somatic antigens O and flagellar antigens H by slide agglutination according to Kauffmann-White scheme (Grimont and Weill, 2007).
The identification scheme requires the initial use of polyvalent antisera that provide a general indication and, subsequently, of specific monovalent antisera against both the somatic antigen O or flagellar antigens H.
Strains were definitively assigned to serovar Typhimurium or S. 4,[5],12:i:– on the basis of the presence or the absence, respectively, of fljB gene tested by PCR (Echeita et al, 2002).
8.3 Antimicrobial susceptibility test
The strains were tested for antimicrobial susceptibility by the disk diffusion method (Clinical and Laboratory Standards Institute, 2006). The antimicrobials tested included the following antibiotic disks (Becton Dickinson)
and concentrations (mg): nalidixic acid (Nx) 30, ampicillin (A) 10, cefotaxime (Ctx) 30, ciprofloxacin (Cp) 5, chloramphenicol (C) 30, gentamicin (G) 10, kanamycin (K) 30, streptomycin (S) 10, sulfonamides (Su) 300, tetracycline (T) 30, trimethoprim–sulphamethoxazole (SXT) 1.25/23.75, according to the Enter-net reference panel (Threlfall et al., 2003). Escherichia coli ATCC 25922 was used as control strain in each experiment.
8.4 Phage typing
Briefly, the technique requires the growth of the isolates on Triptycase Soy Agar (TSA, Oxoid) at 37°C for 18-24 hours. Then some colonies are transferred in 4 ml of Nutrient Broth Double Strength (DSNB, Difco) and incubated for 2 hours at 37°C. At the same time the phages are transferred in microplates to which subsequently is added bacterial suspension, the reaction it is carried out for about 5 minutes and then, with a multichannel pipette, from each well are transferred 7-10 µl of suspension in Nutrient Agar plates (DNA, Difco). The plates inoculated with the suspension phages-bacteria are incubated at 37°C for 18-24 hours. At the end of incubation the reading and interpretation of results is performed, according to the scheme proposed by Phage-typing Reference Laboratory (Health Protection Agency, London, UK).
8.5 Pulsed-field gel electrophoresis (PFGE)
PFGE has been performed in according to Salmgene and Pulse-Net standardized protocol (Peters et al., 2003; Ribot et al., 2006).
Briefly, bacterial strains were streaked on nutrient agar and incubated at 37°C for 18 hours. Cell colonies were resuspended in 1 ml of cell suspension buffer (CSB, 100mM TRIS, 100mM EDTA, pH 8.0) to reach a cell density of 0.38-0.44
It was added proteinase K (0.5 mg/ml final concentration) and subsequently cell suspension was mixed in ratio 1:1 with agarose 2% in TE buffer (10mM TRIS, 1.0mM EDTA pH 8.0), cooled down to 50°C, and dispensed into wells of plug moulds and leaved to set.
When setted, plugs were placed in 2 ml of lysis buffer (50mM Tris, 50mM EDTA, 1% Sarkosyl, 0.1 mg/ml Proteinase K, pH 8.0) and incubated for 2 hours in a shaking waterbath at 55°C. Then the plugs, were washed twice in 5 ml of sterile distilled water, and then washed 3 times in 5 ml TE buffer, at 55°C in a shaking water bath.
A 3 mm of plug has been digested with 50 U Xba-I restriction enzyme (New England, Biolabs, Ipswich, MA) and incubate at 37°C overnight. DNA fragments were separated by using a Bio-Rad CHEF-DR II apparatus, under the following conditions: 6 V/cm, pulse times 2-64 s over 22 hr, temperature 14°C. The gels contained TBE buffer (50mM Tris, 50mM boric acid, 0.5mM EDTA) at 0.5x concentration and 1% agarose. S. enterica serovar Braenderup H9812 was used as the molecular size marker (Hunter et al., 2005).
Dendrogram and cluster analysis were performed using algorithms available within the BioNumerics software package (v. 4.61, Applied Maths, Sint-Martens-Latem, Belgium). The percent similarity between different chromosomal fingerprints was scored by the Dice coefficient. The unweighted pair group method with arithmetic means (UPGMA), with a 1.00% tolerance limit and 1.00% optimization, was used to obtain the dendrogram. DNA profiles differing by one or more DNA fragments were considered as distinct patterns. Strains with a coefficient of similarity ≥80% were considered as closely related genetically. The locally analyzed profiles of the strains were uploaded to the international database established at the Health Protection Agency (HPA; Colindale, London, UK), compared with the profiles in the international database and named, as agreed with the Pulse-Net Europe, with a six letter code followed by a four digit numerical identifier, for example, STYMXB.0006 (Lukinmaa et al., 2006;
8.6 Identification of resistance genes
8.6.1. Total genomic DNA purification
Total DNAs were extracted by Wizard Genomic DNA Purification Kit (Promega, Milan, Italy), by the following procedure: one colony from each strain was inoculated in 3 ml of LB (Luria-Bertani broth), for 18 hours at 37°C. 1 ml of overnight cultures were transferred in a 1.5 ml microcentrifuge tube and centrifuged at 13000 rpm for 2 minutes to pellet the cells. To lyse the cells, the pellets were resuspended in 600 µl of Nuclei Lysis Solution, incubated at 80 °C for 5 minutes and cooled to room temperature. 3 µl of RNase Solution (4 mg/ml) were added to cell lysates and incubated at 37 °C for 15-60 minutes. 200 µl of Protein Precipitation Solution were added to the RNase-treated cell lysates and vortexed vigorously for 20 seconds. The samples were incubated on ice for 5 minutes and then centrifuged at 13000 rpm for 3 minutes. The supernatants containing the DNA were transferred in a clean 1.5 microcentrifuge tube containing 600 µl of room temperature isopropanol. The samples were gently mixed by inversion until the thread-like strands of DNA formed a visible mass. Then, they were centrifuged at 13000 rpm for 2 minutes. The supernatants were carefully removed and 600 µl of room temperature 70% ethanol were added. The samples were centrifuged at 13000 rpm for 2 minutes, the supernatants were carefully removed and the DNAs were dried at 37 °C for 10 minutes. The extracted DNAs were rehydrated by 100 µl of distilled water. These solutions were used for PCR.
8.6.2 Polymerase Chain Reaction Assays (PCR)
Standard PCR amplifications were performed with specific primers for the genes conferring resistance to ampicillin (blaSHV, blaOXA, blaTEM, blaPSE),
streptomycin (strA-strB, aadA2), sulfonamides (sul1, sul2), tetracyclines (tet(A),
tet(B), tet(C), and tet(G), and to detect class 1 integron gene cassettes (Carattoli et
al., 2002; Daly et al., 2000; Gallardo et al., 1999; Miriagou et al., 2004; Pasquali et al., 2004; Randall et al., 2004; Tosini et al., 1998). The reaction was performed with 2.5 U of Taq DNA polymerase (Bioline, London, UK) according to the manufacturer’s recommendations. PCR amplicons of blaTEM gene was fully
sequenced in order to assess which derivatives of TEM gene was harboured.
8.6.3 DNA sequencing
The amplified PCR products of interest and the clones in this study were sequenced by an automatic sequencing Pharmacia Biothec (Bio-Fab Research, Pomezia, Italy). The sequencing primers were synthesized by Primm s.r.l. (Milan, Italy).
The electhropherograms were analyzed by the “Chromas” program (www.tecnelysium.com).
The nucleotide sequence analysis was obtained by NCBI “BLAST” program (www.ncbi.nlm.nih.gov).
8.7 Plasmidic and chromosomal investigation
8.7.1 Plasmid DNA purification
Plasmids were extracted by Qiagen Plasmid Midi Kit (Qiagen, Milan, Italy). In brief, one colony from each strain was inoculated in 25 ml of LB (Luria-Bertani broth), for 18 hours at 37 °C. The overnight cultures were centrifuged at 6000 g for 15 minutes at 4 °C to pellet the cells. The pellets were resuspended in 4 ml of Buffer P1 (50 mM TRIS-HCl, 10 mM EDTA, pH 8.0 containing 100 µg/ml RNasiA). 4 ml of Buffer P2 [200 mM NaOH, 1% SDS (w/v)] were added to the samples, mixed thoroughly by vigorously inverting 4-6 times and incubated at room temperature for 5 minutes. Then 4 ml of chilled Buffer P3 (3.0 M potassium acetate, pH 5.5) were added to the cell lysates, mixed thoroughly by vigorously inverting 4-6 times and incubated on ice for 15 minutes. The samples were centrifuged for 30 minutes at 4 °C. The supernatants containing the plasmids were applied to anion-exchange based columns previously equilibrated by applying 4 ml of Buffer QBT [750 mM NaCl, 50mM MOPS pH 7.0, 15% isopropanol (v/v), 0.15% Triton X-100 (v/v)]. The columns were washed twice with 10 ml of Buffer QC [1.0 M NaCl, 50mM MOPS pH 7.0, 15% isopropanol (v/v)]. The plasmidic DNAs were eluted with 5 ml of Buffer QF [1.25 M NaCl, 50mM MOPS pH 8.5, 15% isopropanol (v/v)]. The plasmidic DNAs were precipitated by adding 3.5 ml of room temperature isopropanol and centrifuged at 15000 g for 30 minutes at 4 °C. The pellets were washed with 2 ml of room temperature 70% ethanol and centrifuged at 15000 g for 10 minutes. The pellets were air-dried and resuspended in 60 µl of distilled water.
8.7.2 Transfer experiments
Conjugation experiments were performed using as recipient strain,
Escherichia coli CSH26NaR. The mating was performed in Nutrient Broth at three range of temperature: 25°C, 28°C and 37°C. Trasconjugants were selected on Luria-Bertani agar plates containing nalidixic acid (40 µg/ml) together with tetracycline (3 µg/ml) or ampicillin (8 µg/ml).
Transformations experiments were performed using DH5α competent cells and transformants selected on LB-agar plates containing ampicillin (50 µg/ml) or tetracycline (8 µg/ml).
8.7.3 PFGE with I-Ceu I
The preparation of genomic DNA was performed as above described, with a few modifications: cell density for plug preparation was 2 at 600 nm, final concentration of proteinase K in the plug was 1 mg/ml, and 1 additional wash both in H2O and in TE buffer. The plugs were digested with 0.04 U of I-Ceu I
enzyme (New England, Biolabs, Ipswich, MA) for 3h at 37 C as described by Liu et al. 1993. DNA fragments were separated on a 0.7% agarose gel and Lambda ladder concatamers (New England, Biolabs, Ipswich, MA) were used as a molecular marker.
8.7.4 Southern blot hybridization experiments
To transfer the digested restriction fragments onto a positively charged nylon membrane, the agarose gel was treated twice for 15 minutes with HCl 0.25M, denaturized in NaOH 0.5M+NaCl 1.5M for 20 minutes, neutralized in Tris-HCl 0.5M pH 7.6 for 20 minutes and finally transferred on a nylon
membrane positively charged in 20X SSC, for 1 night, according the method described by Southern (1975). The DNA was covalently fixed by UV light for 4 minutes. The nylon membranes were used for hybridization experiments by digoxigenin labelled probes for specific resistance genes and for ribosomal DNA. Primers TetBF (5’-ACGTTACTCGATGCCAT-3’) and TetB2 (5’-CCAGTAGCTCCTGTGAT-3’) were used to amplify tet(B) gene probe (EMBL accession no. AM412236). For 16S rDNA and strA-strB were used primers just published (Pezzella et al., 2004; Ziemer and Steadham, 2003).
The membranes were pre-hybridized at 68 °C for 2 hours in a solution containing SSC 5X, N-laurosylsarcosine 0.1% (p/v), blocking solution 1%, SDS 0.02% (p/v) and salmon sperm DNA. Then, the probe (previously boiled for 10 minutes at 100 °C and cooled in ice) was added. Its final concentration was 5-25 ng/ml. The hybridization was at 68 °C for 1 night. Successively, the membrane was washed twice at 68 °C for 5 minutes with SSC 2X + SDS 0.1% and twice at 68 °C for 15 minutes with SSC 0.1X + SDS 0.1%. The DNA fragments recognised by the probe were detected by chemiluminescence, incubating the membrane for 30 minutes with the anti-digoxigenin-alkaline phosphatase diluted 1:10000 in a maleic acid buffer containing blocking solution able to block the not specific sites and detecting the alkaline phosphatase by a specific substrate, the CSPD. The signal was detected by a X-ray film (X-OMAT-AR, Kodak).
8.8 Cosmid library
Genomic DNA was prepared following Ezaki protocol (Ezaki et al., 1988) and partially digested with Sau3A I, as described in Sambrook et al., 2001. SuperCos 1 cosmid vector (Stratagene, M-Medical, Milan, Italy) was digested with XbaI and BamHI, treated with calf intestinal alkaline phosphatase (CIAP), and ligated with genomic DNA. The packaging and titering cosmid packaging reaction has been performed according to Stratagene protocol. Subsequently the
library obtained was transferred onto nylon membrane following the procedures described by Sambrook et al., 2001. Filters obtained were hybridized at the same condition previously described, with specific probes for tet(B) and strA-strB.
The cosmid clones obtained were subsequently subcloned using different restriction enzymes and sequenced, as previously described.
9. Results
9.1 Antimicrobial resistance pattern in Salmonella
Typhimurium and its monophasic variant
The main R-types of the 732 STM and 191 S. 4,[5],12:i:– strains collected through the surveillance system between 2003 and 2006 are reported in table 4. The main R-types are, both in STM and in S. 4,[5],12:i:–, ASSuT and ACSSuT. These 2 R-types showed the same frequency in strains of STM, while S. 4,[5],12:i:– strains showed mainly ASSuT resistance pattern. It is also important to highlight that S. 4,[5],12:i:– strains showed a lower percentage of sensitive strains than STM.
Table 4. Antimicrobial resistance pattern of S. Typhimurium and S. 4,[5],12:i:–
strains. R-type S. Typhimurium n (%) S. 4,[5],12:i:– n (%) ASSuT +/- other 203 (27.7%) 137 (71.7%) ACSSuT +/- other 204 (27.9%) 9 (4.7%) 1-3 Resistances 161 (22%) 27 (14.1%) 4 or + Resistances 42 (5.7%) 11 (5.7%) Susceptible 122 (16.7%) 7 (3.7%) Total 732 191
9.2 PFGE profiles of ASSuT strains
All the 553 STM and S. 4,[5],12:i:– strains showing the ASSuT or ACSSuT patterns, with or without additional resistances, were typeable by PFGE. The results of PFGE analysis for each of profiles containing more than one isolate are shown in Figure 3.
Figure 3: Dendrogram generated by BioNumerics software showing the results of
cluster analysis of 20, non unique, PFGE patterns described among tetra-resistant and penta-resistant S. Typhimurium and S. 4,[5],12:i:– strains. Similarity analysis was performed using the Dice coefficient (Opt: 1.00%-Tol: 1.00%), and clustering was by UPGMA.
Cluster A indicated the tetra-resistant strains and cluster B the penta-resistant strains.
* PFGE profiles of tetra-resistant strains grouping in cluster B. ** PFGE profiles showed by both the R-types.
All the profiles obtained are summarized in Table 5 according to the resistance pattern. The 203 ASSuT STM strains were categorized into 54 different PFGE profiles, 44 of which corresponding to a single isolate and indicated as “other” in Table 5. The predominant profile was STYMXB.0079, which accounted for 53.2% of the strains, followed by STYMXB.0339 (9.8%) and STYMXB.0010 (4.4%).
The 137 ASSuT S. 4,[5],12:i:– strains belonged to 17 different profiles, 13 of which corresponded to a single isolate. The STYMXB.0079 profile was prevalent also in this group of strains (73%), followed as well by STYMXB.0010 (9.5%) and STYMXB.0339 (5.8%).
The coefficients of similarity (F) of the predominant STYMXB.0079 with the STYMXB.0339 and STYMXB.0010 profiles were 0.84 and 0.91, respectively (Figure 3). The STYMXB.0079 and STYMXB.0010 profiles were also observed among the 9 ACSSuT S. 4,[5],12:i:– strains examined.
Among the 204 ACSSuT STM strains, 41 PFGE profiles were observed, 31 of which were unique. STYMXB.0061 and STYMXB.0067, which showed an F value of 0.95, were the prevalent PFGE profiles observed, accounting for 67.1% of strains.
Table 5. Distribution of PFGE profiles among tetra-R and penta-R
S. Typhimurium and S. 4,[5],12:i:– strains.
* Cluster A and B were defined by Bionumerics cluster analysis as shown in Figure 3. PFGE Profile (Cluster*) S. Typhimurium S. 4,[5],12:i:– ASSuT +/- other ACSSuT +/-other ASSuT +/- other ACSSuT +/- other STMXB.0079 (A) 108 (53.2) 5 (2.5) 100 (73) 3 (33.3) STMXB.0339 (A) 20 (9.8) - 8 (5.8) - STMXB.0010 (A) 9(4.4) - 13 (9.5) 1(11.1) STMXB.0132 (A) 5 (2.5) - - - STMXB.0022 (A) 4 (2) - - - STMXB.0083 (A) 4 (2) - - - STMXB.0080 (A) 1 (0.5) - 3 (2.2) 1 (11.1) STMXB.0061 (B) - 76 (37.2) - 1(11.1) STMXB.0067 (B) - 61 (29.9) - - STMXB.0114 (B) - 6 (2.9) - - STMXB.0179 (B) - 5 (2.4) - - STMXB.0020 (B) - 4 (2.0) - - STMXB.0015 (B) - 4 (2.0) - - STMXB.0260 (B) - 3 (1.5) - - STMXB.0013 (B) - 2 (1.0) - - STMXB.0115 (B) - 2 (1.0) - - STMXB.0233 (B) - 2 (1.0) - - STMXB.0058 (B) 3 (1.5) 3 (1.5) - - STMXB.0086 (B) 3 (1.5) - - - STMXB.0051 (B) 2 (0.9) - - - Other profiles 44 (21.7) 31(15.1) 13 (9.5) 3 (33.3) Total 203 (100.0) 204 (100.0) 137 (100.0) 9 (100.0)
Overall, two large clusters with a 73% of genetic homology were identified. Cluster A included 275 out of the 283 (97.2%) ASSuT strains, while cluster B, included 169 out of the 179 (94.4%) ACSSuT strains, regardless of STM or S. 4,[5],12:i:– serovar. A close genetic relationship (F > 0.80 ) was observed among the strains belonging to each cluster. The few exceptions included 8 tetra-resistant strains with PFGE profiles (STYMXB.0086, STYMXB.0051 and STYMXB.0058) grouping in cluster B, and 10 penta-resistant strains with profiles grouping in cluster A (STYMXB.0079, STYMXB.0010 and STYMXB.0080).
9.3 Phage typing of ASSuT strains
Phage typing was performed for 357 STM and 69 S. 4,[5],12:i:–, including tetra-resistant and penta-resistant strains. The ASSuT strains were mainly not typeable with this technique (DTNT) or belonged to phage type U302: 23% and 22.3% of STM strains, 46% and 27.9% of S. 4,[5],12:i:– strains, respectively. Most of the ACSSuT STM strains (70.2%) belonged to DT104, as expected, while none of the ACSSuT S. 4,[5],12:i:– belonged to this phage type.
The associations between PFGE profiles and phage types are described in detail in Tables 6 and 7.
Table 6: Association between PFGE profile and phage types of the 357 S. Typhimurium strains according to resistance pattern.
Phagetype
PFGE profile
ASSuT +/- other ACSSuT +/- other
STMXB.0079 STMXB.0339 STMXB.0010 Other STMXB.0061 STMXB.0067 STMXB.0079 Other DTNT* 27 3 - 9 2 - 2 5 U302 27 1 1 9 4 4 2 5 DT7 15 - - 6 - - - - RDNC** 3 2 - 9 - - - 7 DT120 7 - 6 9 2 2 - 11 DT20 3 - - - - DT194 2 - - 2 - - - - DT104 - - - 4 54 45 - 32 DT193 - - - 8 - - - - DT22 - 7 - - - - Other 1 3 - 6 - - - 10 Total 85 16 7 62 62 51 4 70 *. DTNT: Unable to be typed
Table 7: Association between PFGE patterns and phage types of the 69 strains of S. 4,[5],12:i:– according to resistance pattern.
Phagetype
PFGE profile
ASSuT +/- other ACSSuT +/- other
STMXB.0079 Other STMXB.0010 STMXB.0079 STMXB.0080 Other U302 16 1 - 1 - 3 DTNT* 22 6 1 1 1 - DT7 1 1 - - - - RDNC** 4 - - - - - DT120 1 5 - - - - Other 1 3 - - - 1 Total 45 16 1 2 1 4 *. DTNT: Unable to be typed
9.4 Identification and localization of resistance genes
By using the Pulse-Net Europe database (Lukinmaa et al., 2006), we were able to identify ASSuT strains, both STM and S. 4,[5],12:i:–, isolated in other European countries, such as Denmark and UK, with the same PFGE patterns as the Italian strains, suggesting that the ASSuT clone is circulating in different European countries. A unique exception is represented by STMXB.0131, a PFGE profile not shown by Italian strains, but showing a 79.8 % of homology with the PFGE profiles typical of ASSuT strains.A total of 64 STM and S. 4,[5],12:i:– strains, isolated in Italy, Denmark and United Kingdom, with the ASSuT resistance pattern and a genetic similarity ≥ 79.8% by PFGE, were selected in order to identify their resistance genes by PCR. In addition, 11 STM strains showed different R-types but which were both genetically related (8 strains) and unrelated (3 strains) to ASSuT strains, were also included for comparison. All strains were isolated from human infections in Italy (38 strains), Denmark (29 strains) and UK (8 strains). A detailed description of the strains is reported in Table 8.
Table 8: Summary of the 75 S. Typhimurium and S. 4,[5],12:i:– strains included in this study. Dendrogram was generated by
BioNumerics software. Similarity analysis was performed using the Dice coefficient (Opt: 1.00%-Tol: 1.00%), and clustering was by UPGMA.
PFGE, pulsed-field gel electrophoresis; IT, Italy; UK, United Kingdom; DEN, Denmark; Antibiogram designations: A, ampicillin; S, streptomycin; Su, sulfonamides; T, tetracyclines
The strains were tested by PCR amplification for the presence of the following resistance genes: blaSHV, blaOXA, blaTEM,blaPSE, strA-strB, aadA2, sul1,
sul2, tet(A), tet(B), tet(C), and tet(G). Strains were also tested for the presence of
class 1 integron. The results of PCR analyses are shown in Table 9.
All 64 ASSuT strains, were positive for blaTEM-1, strA-strB, sul2 and tet(B)
and negative for all the other genes tested and for class 1 integron(s). The same resistance genes were found in all strains of R-type ASSu and two of R-type SSuT strains, in accordance with their resistance profiles.
The strains not-ASSuT positive for tet(B) were genetically related to ASSuT strains: four strains with R-types SSuT or T exhibited the PFGE profile STYMXB.0079, and one strain resistant only to tetracyclines showed the PFGE profile STYMXB.0131, a related profile observed only in some Danish ASSuT strains. PFGE profiles STYMXB.0079 and STYMXB.0083 were also showed by three strains of R-type ASSu, which, with the exception of tet(B), showed the same resistance gene profile of the ASSuT strains, (Table 9). Regarding the tetracycline resistance genes different from tet(B), tet(A) was found in 2 strains with SSuT and T resistance patterns, and tet(C) in one tetracycline-resistant strain isolated in Italy.
Table 9: Antimicrobial resistance genes identified among S. Typhimurium
and S. 4,[5],12:i:– strains
N° of strains
R-type PFGE-type tet(A) tet(B) tet(C) strA- strB sul2 tem 38a ASSuT STYMXB.0079 - + - + + + 4 ASSuT STYMXB.0339 - + - + + + 5b ASSuT STYMXB.0010 - + - + + + 12 ASSuT STYMXB.0083 - + - + + + 3 ASSuT STYMXB.0022 - + - + + + 2 ASSuT STYMXB.0131 - + - + + + 2 SSuT STYMXB.0079 - + - - + - 2 ASSu STYMXB.0079 - - - + + + 1 ASSu STYMXB.0083 - - - + + + 2 T STYMXB.0079 - + - - - - 1 T STYMXB.0131 - + - - - - 1 T STYMXB.0089 - - + - - - 1 T STYMXB.0103 + - - - - - 1 SSuT STYMXB.0336 + - - + + -
a: 10 of these strains belong to S. 4,[5],12:i:– b: 2 of these strains belong to S. 4,[5],12:i:–
Conjugation and transformation experiments were performed on two strains of R-type ASSuT, belonging to 2 different PFGE profiles, STYMXB.0079 and STYMXB.0339. Both the experiments failed, suggesting inability of horizontal transfer and a chromosomal localization of the resistance genes.
Chromosomal localization of resistance genes was investigated by PFGE using the I-Ceu I restriction enzyme on a selection of 41 strains, including 18 strains from Italy, 15 from Denmark and 8 from UK. The strains were representative of each PFGE profile and all harbouring tet(B) gene. Three danish strains of R-type ASSu were included as well. Chromosomal digestion yielded seven PFGE fragments with sizes from 40 kb to greater than 3,000 kb. Southern blot hybridization was performed with specific probes for tet(B), strA-strB and 16S r-DNA. The probes specific for the resistance genes bound to an approximately 750 kb fragment, confirming the chromosomal location of the resistance determinants (Figure 4).
Figure 4: (A) Pulsed field gel electrophoresis with I-Ceu I on a subset of
strains with the following features. Lanes 1, ASSuT STYMXB.0079; Lanes 2, ASSuT STYMXB.0339; Lanes 3, ASSuT STYMXB.0010; Lanes 4, ASSuT STYMXB.0083; Lanes 5, ASSuT STYMXB.0131; Lanes 6, ASSu STYMXB.0079; Lanes 7, ASSu STYMXB.0083; Lanes 8, T STYMXB.0079; Lanes 9, T STYMXB.0131. Molecular weight marker (Lambda ladder concatamers) is in lane M. (B to D): Southern blot hybridization was performed with tet(B), strA-strB and DNA16S probes.
9.5 Genetic characterization of chromosomal resistance
island ASSuT
The characterization of the chromosomal resistance region containing the
tet(B), strA-strB, sul2 and blaTEM-1 genes was performed by construction of a
genomic library of a strain ASSuT, PFGE profile STYMXB.0079. The hybridization of the colonies with the tet(B), strA-strB specific probes carried to the picking of 2 clones containing two overlapping inserts of above 36-kb and 34-kb respectively. These two inserts were sequenced and the sequence analysis leaded to the assembly of 34391-bp region conferring the ASSuT resistance. The 34 kb genomic region contains 2 resistance island (RI) inserted in two adjacent loci of the chromosome, involved in the transport and metabolism of sugar (Figure 5). In particular the RI1 island, conferring resistance to ampicillin, streptomycin and sulfonamides, was inserted at the STM2753 gene causing a deletion of 190 bp, while the RI2 island, harbouring tet(B) gene, was inserted at the gene STM2759, causing a deletion of 6 bp. The chromosomal fragment comprised between the 2 RI encodes genes from STM2753 to STM2759 but in inverted orientation (Figure 5).
![Table 4. Antimicrobial resistance pattern of S. Typhimurium and S. 4,[5],12:i:–](https://thumb-eu.123doks.com/thumbv2/123dokorg/8052090.123265/34.892.167.765.659.912/table-antimicrobial-resistance-pattern-s-typhimurium-s-i.webp)



![Table 7: Association between PFGE patterns and phage types of the 69 strains of S. 4,[5],12:i:– according to resistance pattern](https://thumb-eu.123doks.com/thumbv2/123dokorg/8052090.123265/40.1263.141.1097.238.553/table-association-pfge-patterns-strains-according-resistance-pattern.webp)