ALMA MATER STUDIORUM
UNIVERSITA' DI BOLOGNA
FACOLTA' DI SCIENZE MATEMATICHE FISICHE E NATURALI
Corso di laurea magistrale in BIOLOGIA MARINA
Molecular phylogenesis of Mediterranean Octocorals
Relatore:
Presentata da:
Prof. Abbiati Marco
Marchiselli Simone
Correlatore:
Dott.ssa Costantini Federica
(III sessione)
21-Feb-2013
Index
1. Introduction pp. 1
1.1. Ecosystem functions and treats of Mediterranean coralligenous pp. 1 1.2. Mediterranean octocorals morphological characteristics and classification pp. 3 1.2.1 Mediterranean octocorals distribution pp. 10
1.3. Octocorals phylogenetic analyses pp. 12
1.4. Mediterranean Octocorallia dated phylogeny pp. 14
1.4.1 The red coral pp. 16
1.5 Aims pp. 18
2. Materials and methods pp. 19
2.1. Sampling pp. 19
2.2. DNA extraction, amplification and sequencing pp. 22
2.3. Data analyses pp. 23
2.4. Alignment methods pp. 24
2.5. Substitution model test and gene partition pp. 25
2.6. Maximum likelihood analyses pp. 26
2.7. Bayesian analyses pp. 27
2.8. Estimation of divergence time pp. 29
3. Results pp. 31
3.1. Mediterranean MtMSH genetic variability pp. 31
3.2. Mediterranean 16S genetic variability pp. 38
3.3. Mediterranean phylogenetic analysis pp. 43
3.3.1 MtMSH Mediterranean trees pp. 43
3.3.2. 16S Mediterranean trees pp. 46
3.4. Mediterranean and Atlanto-Pacific species phylogenetic analysis pp. 48
3.4.1. MtMSH trees pp. 48
3.4.2. 16S trees pp. 52
3.4.3 MtMSH + 16S combined trees pp. 55
3.5. Divergence time pp. 55
3.5.1 Fix mutation rate estimate pp. 55
3.5.2 Calibrated point estimation pp. 59
4. Discussion pp. 62
4.1 The Mediterranean Octocorals phylogeny pp. 62
4.2 The Mediterranean octocorals origin pp. 68
4.3 Conclusion pp. 72
1. Introduction
1.1. Ecosystem functions and treats of Mediterranean coralligenous
The so-called "coralligène" (Ballesteros 2006) is one of the main Mediterranean hard substratum habitat, which generally occur between 20 and 120 m depth along coasts (Laborel 1987) and seamounts (Clark et al. 2010). The name “coralligenous” could be originate from the findings of red coral branches and calcareous organisms in trawling hauls of semi dark sub littoral bottoms with coarse gravel (Tsounis et al. 2010; Ballesteros 2006). These habitats as the tropical reefs are considered a species diversity ‘‘hot spot’’; therefore, these reefs are intrinsically valuable for their biological diversity and for the ecological processes that they can support (Ballesteros 2006; Coll et al. 2010). However, we are still only beginning to understand the principal ecological aspects of coralligenous assemblages, including the environmental factors (temperature, salinity, nutrition) and biological processes (reproductive biology, molecular genetics, predation, parasitism and bioerosion), which regulate their life and distribution (UNEP 2004; UNEP 2010).Several sources of human disturbance threaten Mediterranean coralligenous assemblages, e.g.: pollution, sediment smothering and deposition, bottom-trawling, deep-sea mining, hydrocarbon extraction, waste disposal, recreational fishing and diving (Airoldi & Beck 2007; Airoldi et al. 2008; see for review). Moreover, environmental changes, leading to mass mortality events (Cerrano et al. 2000), invasions by alien species (Occhipinti-Ambrogi 2007) and acidification (Ramirez-Llodra et al. 2011), are additional sources of disturbance to these habitats. Other causes of mortality for these benthic assemblages are bottom hypoxia/anoxia events and suffocation by mucilaginous aggregates (Ponti et al. 2011 and references therein). Due
to the above mentioned features, the Mediterranean coralligenous should be a priority for several international bodies (e.g. UNEP, IUCN, EU organizations; Fraschetti et al. 2011). In order to support the protection of coralligenous reefs, identification of species cladogenesis and preservation of evolutionarily distinct lineages (as for example endemic species) are important aspects to take in account for their conservation (Fukami et al. 2004).
Among the coralligenous habitat, octocorals (Anthozoa: Octocorallia) are one of the main taxonomic groups with a high number of species. Moreover, some of them have a pivotal role as ecosystem engineers. In fact, they act as refuge or nursery zone, may reduce current flow velocity, can stabilise soft substrata and can experience a sort of ‘‘buffer zone’’ where environmental modifications occur slower (Cerrano et al. 2009). Moreover, coral ecosystems also support fisheries (D’Onghia et al. 2011; Soeffker et al. 2011) and have been identified as important sources of marine natural products (Leal et al. 2012). Treats as fishing impacts on octocorals are less well known (Stone, 2006; Edinger et al., 2007). The resilience of such communities is perceived to be very low as some octocorals have slow growth rates and high longevity (Freiwald et al., 2004; Althaus et al., 2009). A significant problem with the identification of vurnerable marine ecosystems (VMEs), such as octocoral habitat, is the lack of information of benthic communities throughout much of the deep sea (Yesson et al 2012).
1.2. Mediterranean octocorals morphological characteristics and
classification
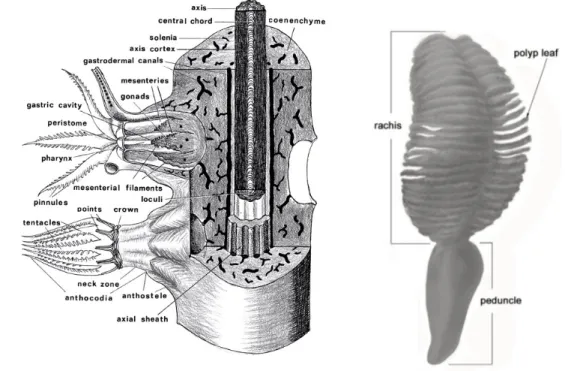
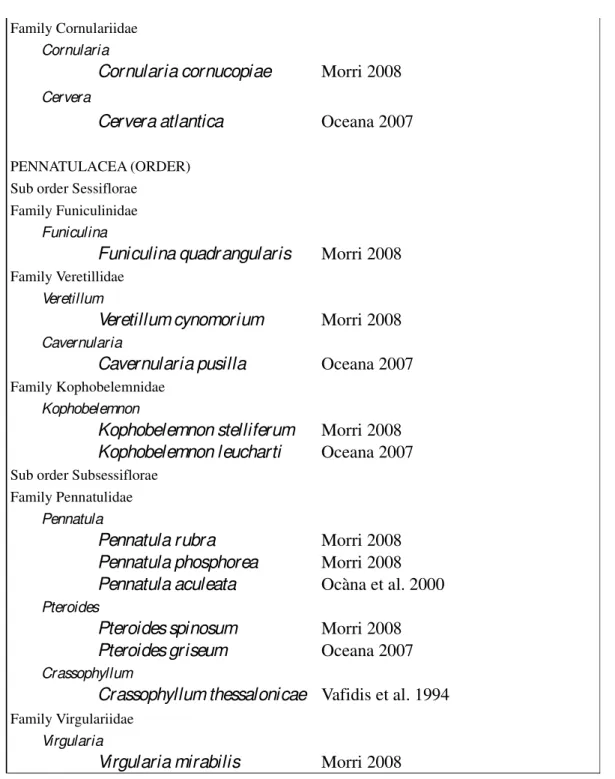
Within Anthozoa, the subclass Octocorallia included approximately 3,000 existing species (Daly et al. 2007), the diagnostic apomorphies (an evolutionary trait that is unique to a particular species and all its descendants; Dictionary of Biology 2000) are the eight tentacles and the eight mesenteries of polyps (Daly et al. 2007; Figure 1.1), as suggested by the group name.
The current classification system divides Octocorallia into three orders: Alcyonacea, Pennatulacea, and
Helioporacea (Bayer 1981). Only the first two orders live in the Mediterranean Sea. The primary morphological characters that have been used to define the Alcyonacea families are the overall growth forms of colonies, details of the composition of the skeletal axis (if present is characterized by having some form of hard skeletal structure composed of some combination of calcium carbonate and the horn-like, proteinaceous material gorgonin; McFadden et al. 2006) and the shape or the arrangement of sclerites (free skeletal elements embedded in the tissue of polyps and coenenchyme; Daly et al. 2007). The presence of pinnules (lateral extensions) on the tentacles is also considered
diagnostic, although this character is absent in several taxa (Daly et al. 2007; Figure 1.1). Differently, the Pennatulacea (sea pens) are recognizable by the unique colony form, in which the single axial polyp differentiates into a proximal peduncle and a distal rachis. This represents a morphological synapomorphy (i.e. the possession of apomorphic features by two or more taxa, which related the group evolutionarily; Dictionary of Earth Science 1999; Figure 1.1) that clearly unites species from the order Pennatulacea and distinguishes its members from all other octocorals (Daly et al. 2007).
Figure 1.1. On the left, scheme of a gorgonacean ctocoral (from Bayer et al. 1983). On right, picture of a Pennatula sp. of Subsessiflorae sub-order (Williams et al. 2011).
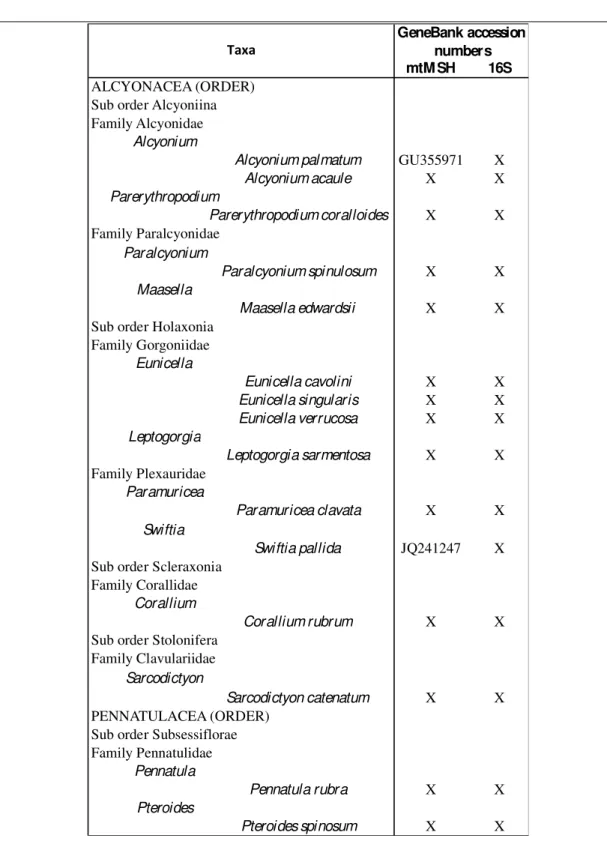
Within Anthozoa, the subclass Octocorallia included approximately 3,000 existing species (Daly et al. 2007). In the Mediterranean Sea, diverse scientific sources such as S.I.B.M. Italian species checklist (Morri 2008), or Oceana “Mediterranean corals” reports (Oceana 2007) and several zoological papers recognized 56 Octocorallia species to date (Table 1.1).
Papers references
OTTOCORALLIA (SUBCLASS) ALCYONACEA (ORDER) Sub order Alcyoniina Family Alcyonidae
Alcyonium
Alcyonium palmatum Morri 2008
Alcyonium acaule Morri 2008
Parerythropodium
Parerythropodium coralloides Ocàna et al. 2000
Family Paralcyonidae
Paralcyonium
Paralcyonium spinulosum Morri 2008
Maasella
Maasella edwardsii Morri 2008
Family Nidaiidae
Nidalia
Nidalia studeri Lòpez-Gonzales 2012
Sub order Calcaxonia Family Ellisellidae
Ellisella
Ellisella paraplexauroides Vafidis et al. 1994
Viminella
Viminella flagellum Giusti et al. 2011
Family Isididae
Isidella
Isidella elongata Morri 2008
Family Primnoidae
Callogorgia
Callogorgia verticillata Morri 2008
Family Dendrobrachiidae
Dendrobrachia
Dendrobrachia bonsai Sartoretto 2012
Dendrobrachia fallax Zibrowius & Taviani (2005)
Sub order Holaxonia Family Acanthogorgiidae
Acanthogorgia
Acanthogorgia hirsuta Morri 2008
Acanthogorgia armata Ocàna et al. 2000
Family Gorgoniidae
Eunicella
Eunicella cavolini Morri 2008
Eunicella singularis Morri 2008
Eunicella verrucosa Morri 2008
Eunicella filiformis Gori et al. 2012 and references therein
Eunicella gazella Gori et al. 2012 and references therein
Eunicella labiata Gori et al. 2012 and references therein
Filigorgia
Filigorgia guineensis Ocàna et al. 2000
Leptogorgia
Leptogorgia sarmentosa Morri 2008
Leptogorgia viminalis Oceana 2007
Family Plexauridae
Bebryce
Bebryce mollis Morri 2008
Muriceides
Muriceides lepida Morri 2008
Paramuricea
Paramuricea clavata Morri 2008
Paramuricea macrospina Morri 2008
Placogorgia
Placogorgia coronata Vafidis et al. 1994
Placogorgia massiliensis Vafidis et al. 1994
Spinimuricea
Spinimuricea klavereni Morri 2008
Spinimuricea atlantica Vafidis et al. 1994
Swiftia
Swiftia pallida Morri 2008
Swiftia dubi a Vafidis et al. 1994
Villogorgia
Villogorgia bebrycoides Morri 2008
Sub order Scleraxonia Family Corallidae
Corallium
Corallium rubrum Morri 2008
Sub order Stolonifera Family Clavulariidae
Clavularia
Clavularia crassa Morri 2008
Clavularia marioni Morri 2008
Clavularia carpediem Oceana 2007
Rolandia
Rolandia coralloides Morri 2008
Sarcodictyon
Sarcodictyon catenatum Ocàna et al. 2000
Scleranthelia
Scleranthelia rugosa Oceana 2007
Family Cornulariidae Cornularia
Cornularia cornucopiae Morri 2008
Cervera
Cervera atlantica Oceana 2007 PENNATULACEA (ORDER)
Sub order Sessiflorae Family Funiculinidae
Funiculina
Funiculina quadr angularis Morri 2008 Family Veretillidae
Veretillum
Veretillum cynomorium Morri 2008
Cavernularia
Cavernularia pusilla Oceana 2007 Family Kophobelemnidae
Kophobelemnon
Kophobelemnon stelliferum Morri 2008
Kophobelemnon leucharti Oceana 2007 Sub order Subsessiflorae
Family Pennatulidae Pennatula
Pennatula rubra Morri 2008
Pennatula phosphorea Morri 2008
Pennatula aculeata Ocàna et al. 2000
Pteroides
Pteroides spinosum Morri 2008
Pteroides griseum Oceana 2007
Crassophyllum
Crassophyllum thessalonicae Vafidis et al. 1994
Family Virgulariidae Virgularia
Virgularia mirabilis Morri 2008
Table 1.1. List of Mediterranean octocorals. Species classification was checked by the world register of marine species (WoRMS) and the European marine register of biodiversity and ecosystem functions (MaRBEF).
The Mediterranean Alcyonacea can be divided into five sub-ordinal groups (Alcyoniina, Calcaxonia, Scleraxonia, Holaxonia and Stolonifera; Daly et al. 2007). Alcyoniina comprise two families of soft corals (Alcyonidae and Paralcyonidae) in which polyps are embedded within a fleshy mass of coenenchymal tissue (McFadden
et al. 2010; Figure 1.2-H, I, M, N). Only three Calcaxonia families, characterized by a solid axial skeleton, live in the Mediterranean Sea and most species are distributed in deep water (Watling et al. 2011). In the Scleraxonia sub-order the axis is composed largely of hard structure (McFadden et al. 2010). Just one species of this sub-order is represented in the Mediterranean: Corallium rubrum (Figure 1.2-D). The major number of Mediterranean octocorals is represented by the Holaxonia, the typical sea fan or sea whips with soft proteinaceous skeletal. Many families of this group are defined by skeletal axis details, such as the diameter of the hollow central core, and the presence of scleritic or non-scleritic calcite (McFadden et al. 2010; Figure 1.2-A, B, C, E, F and G). Stolonifera includes two families (Clavularidae and Cornularidae; Figure 1.2-L) in which polyps are connected basally by stolons or thin membranes (McFadden et al. 2010). Pennatulacea are distinguished in two sub order by the conspicuous presence of polyp leaves, often strap-shaped wing-like expansions, which can contain numerous (sub-order Subsessiflorae; Figure 1.2-O, P) or less (sub-order Sessiflorae; Williams 2011). Two families of leaf sea pens (Virgularidae and Pennatulidae) and three of Sessiflorae (Funiculinidae, Kophobelemnidae and Veretillidae) were found in the Mediterranean Sea to date.
Figure 1.2. Picture of some Mediterranean Octocorals: Leptogorgia sarmentosa (A),
Swiftia pallida (B), Paramuricea clavata (C), Corallium rubrum (D), Eunicella
verrucosa (E), Eunicella singularis (F) Eunicella cavolini (G), Paralcyonium
spinulosum (H), Maasella edwardsii (I), Sarcodictyon catenatum (L),
Parerythropodium coralloides (M), Alcyonium sp. (N), Pteroides spinosum (O) and
1.2.1 Mediterranean octocorals distribution
The number of endemic species in the Mediterranean Sea are high, averaging more than one quarter of the whole Mediterranean biota. Compared to world estimates, the Mediterranean apparently harbours somewhat between 4% and 18% of the world marine species, with large differences according to the group considered (Bianchi and Morri 2000). Vicariance (Cunningham & Collins 1998), historical environmental factors related to habitat, currents and glaciations (Roy et al. 1996; Wares 2002) combined with biological species-specific traits such as reproduction (Jackson 2001), play a pivotal role in shaping the pattern of inter- and intra-specific differentiation of many species inhabiting the Mediterranean Sea (see Paternello et al. 2007 and references therein). In fact, the biogeography of this Sea strongly reflects both ancestral and modern discontinuities in marine environment. (Longhurst 1998). For example, Octocorals are important members of a wide variety of marine communities, from shallow tropical coral reefs to the deep sea bottoms and cliffs (McFadden et al. 2006). But their species diversity within Mediterranean Sea is constrained by some factors.
However, among the Mediterranean octocorals species selected for this thesis, only Eunicella cavolini two species were was recognized as true endemism,
Maasella edwardsii, and Eunicella cavolin(i (Rossi 1959; Table 1.2). Some other species as Corallium rubrum (Zibrowius et al. 1984), Maasella edwardsii (Parenzan 1977) and Eunicella singularis (Rossi 1959) were largely present and typical of the Mediterranean Sea, but they have been found also along the South-Eastern coasts of the Atlantic Ocean. The rest of the analysed species, such as Paramurice clavata and
Pennatula rubra, had a widespread distribution generally extended to all the North-Est Atlantic, but with few scientific observation.
OTTOCORALLIA (SUB CLASS) ALCYONACEA (ORDER) SUB ORDER ALCYONIINA Family Alcyonidae
Alcyonium
Alcyonium palmatum NE-Atlantic, Mediterranean
Alcyonium acaule NE-Atlantic, Mediterranean
Parerythropodium
Parerythropodium coralloides Wester Mediterranean, N-Europe
Family Paralcyonidae
Paralcyonium
Paralcyonium spinulosum NE-Atlantic, Mediterranean
Maasella
Maasella edwardsii Mediterranean
SUB ORDER HOLAXONIA Family Gorgoniidae
Eunicella
Eunicella cavolini Mediterranean
Eunicella singularis Mediterranean
Eunicella verrucosa N-Atlantic, Mediterranean
Leptogorgia
Leptogorgia sarmentosa NE-Atlantic, Mediterranean
Family Plexauridae
Paramuricea
Paramuricea clavata N-Atlantic, Mediterranean
Swiftia
Swiftia pallida N-Atlantic, Mediterranean
SUB ORDER SCLERAXONIA Family Corallidae
Corallium
Corallium rubrum Mediterranean, N-Africa Atlantic coast
SUB ORDER STOLONIFERA Family Clavulariidae
Sarcodictyon
Sarcodictyon catenatum NE-Atlantic, North Sea , Mediterranean
PENNATULACEA (ORDER) SUB ORDER SUBSESSIFLORAE Family Pennatulidae
Pennatula
Pennatula rubra E-Atlantic, Mediterranean
Pteroides
Pteroides spinosum E-Atlantic, Mediterranean, NE-Indian
Taxa Species distribution
Table 1.2. Geographical distribution of the species analysed in the present thesis.
1.3. Octocorals phylogenetic analyses
Molecular tools are extremely useful to increase the knowledge on classical taxonomy based just on morphological traits, by comparing the morphological differences among species with their genetic features. Moreover they permit to figure out the evolutionary relationship among species, place them within their evolutionary historical framework and understand the relative timing of lineage splitting (Palumbi 2012). These reconstructions also depend on understanding the temporal framework of species, using the lens of paleontological dating, climate records, human historical records or other links to the past (Palumbi 2012).
Mitochondrial DNA loci represent useful molecular markers for genetic analysis, principally due to features as feasibility, relatively simple genetic structure and straightforward mode of genetic transmission (Avise et al. 1987). The France et al. (1996) work on 16S rDNA revealed little variation among families of octocorals, having values of sequence divergence ranging from 2.7 to 6.3%, compared to those observed among families of Hexacorallia (16.1–26.3%; McFadden et al. 2010). Possible explanations for the reduced rates of divergence observed seems due to the presence of a gene (mtMSH), which may code for a mitochondrial DNA mismatch-repair system (France and Hoover 2002). In fact, nearly 16% of the octocoral mtDNA sequence is occupied by a novel open reading frame (ORF hereafter) that has not been found in any mitochondrial metazoans genome than the Octocorallia group up to now (France and Hoover 2002; McFadden et al. 2010, Bilewitch and Degnan 2011). Based on the octocoral translated sequence 19.7% similarity to the yeast nuclear Mutation Suppressor Homolog 1 (MSH1) gene, it was suggested that the novel ORF might be active in DNA mismatch repair. Accordingly, it was first termed ‘mtMSH’ as a mitochondrial homolog of the eukaryotic MSH family (Bilewitch and Degnan 2011).
The origin of this gene in the Octocorallia mitochondrial genome has recently been attributed to a horizontal gene transfer from a non-eukaryotic source (Bilewitch and Degnan 2011; Park et al. 2012). The mtMSH gene is approximately twice more polymorphic compared to most other protein-coding regions in the octocoral mitochondrial genome (France and Hoover 2001). It has been widely used for genus-level and species-genus-level phylogenetic studies, but generally has not resulted in better discrimination of congeneric morphospecies (McFadden et al. 2010 and references therein).
Phylogenetic studies obtained using 18S rDNA or 16S mtDNA sequences have supported the monophyletic origin of Octocorallia (Bertson et al. 1999, Sánchez et al. 2003a). Nevertheless, many phylogenetic studies on Octocorallia reveal that traditional taxonomic rank doesn't mirror its molecular phylogenetic structure (McFadden et al. 2010 and references therein). At lower taxonomic levels (family/genus), phylogenetic analyses shown that the classification of species was congruent with taxonomy (Sánchez et al., 2003b; Wirshing et al., 2005; McFadden et al. 2006). Contrarily, at higher levels, the current order/family ranks of taxonomy were not supported by molecular data (McFadden et al. 2006) and reflect grades of morphological construction rather than evolutionary clades (Daly et al. 2007).
The most complete phylogeny of Octocorallia published to date (McFadden et al. 2006; McFadden et al. 2010) sequenced two portions of the mitochondrial protein-coding regions (MSH and ND2) of 115 genera representing 29 of 46 families. Results shown that octocorals were split in three clades:
• Clade 1 (Holaxonia-Alcyoniina): included all members of Holaxonia, the majority of Alcyoniina, and some representatives of Scleraxonia and Stolonifera;
• Clade 2 (Calcaxonia-Pennatulacea): comprised Calcaxonia plus Pennatulacea;
• Clade 3 (Anthomastus-Corallium): a small clade including only precious coral genus Corallium and several taxa of Alcyoniina (McFadden et al. 2010).
Furthermore few taxa of Scleraxonia and Stolonifera occupy unresolved positions near the base of the phylogeny (McFadden et al. 2006). Complete mtDNA genomes studies based on few species (Park et al. 2012) continue to support the two major clades of Octocorallia (Holaxonia–Alcyoniina and Calcaxonia– Pennatulacea) of McFadden et al. (2006).
Despite the increasing amount of molecular information about octocorals (see McFadden et al. 2010 for a review), there are no phylogenetic studies on Mediterrenean species.
1.4. Mediterranean Octocorallia dated phylogeny
The geological history of the Mediterranean Sea has provided sample opportunities for allopatric speciation to the Atlantic species (McFadden et al. 1999). Around 6 millions years ago (MYA hereafter) the connectivity between Mediterranean Sea and any other ocean was lost resulting in the biologically critical Messinian Salinity Crisis (MSC hereafter), during which the vast majority of the Mediterranean Sea dried out (Krijgsman et al. 1999; Donald et al. 2011). The MSC ended at the start of the Pliocene (5.3 Ma) with the Zanclean Flood and the reconnection of the Mediterranean to the Atlantic Ocean via the Strait of Gibraltar (Harzhauser et al., 2007). Many molecular studies analysed biogeographical separation between the Atlantic and Mediterranean biota (see review Paternello et al. 2007, Meynard et al.
2012, Donald et al. 2011) suggesting that the present-day biota is largely the result of colonization, mostly from the Atlantic Ocean after the MSC (Patarnello et al. 2007).
The fossil record has traditionally provided the way to date events in the history of life (Wray 2001).. The anthozoans records were biased and incomplete, since many species lack any solid skeleton, which would be preserved more readily (Berntson 1998)., Information on fossil species principally includes two Paleozoic octocorals documentations up to now, one from the lower Ordovician (471–478 Ma; Cope 2005) and one from the lower Silurian (425–435 Ma; Bengtson 1981). The Ordovician fossil is the earliest gorgonian coral known belonging to the order Alcyonacea. The Silurian fossil had been the earliest representative of the order Alcyonacea known before the Cope (2005) work. Moreover, regarding the genus
Corallium, Vertino et al. (2010) suggested that populations of C. rubrum have been inhabited the Mediterranean basin without interruptions from the early Pleistocene (more than 2 MYA).
Like the fossil record, the genomic record can provide a valuable source of information regarding the timing of evolutionary events whether correctly interpreted (Wray 2001). Estimating divergence time involves calibrating the rate at which protein or DNA sequences evolve, and then estimating when two evolutionary lineages diverged, using the sequences’ differences among their living representatives (Wray 2001). A dated phylogeny on Mediterranean Octocorallia and their closely related (cogeners) oceanic species could be useful to identify the evolutionary origin of Mediterranean corals and to test whether recent Mediterranean geological events (such as MSC) were the main force that drove octocorals speciation within the basin.
1.4.1 The red coral
Corallidae family comprises nearly 32 described species which are found throughout the world in tropical, subtropical and temperate oceans. Seven of them live in the Atlantic Ocean: Corallium niobe, Corallium maderense, Corallium medea,
Corallium johnsoni, Corallium tricolor, Corallium bathyrubrum and Corallium bayeri (Watling and Auster 2005, Simpson and Watling 2011). All the other species inhabit the Pacific and Indian Ocean, excepted the just one living in the Mediterranean Sea: the red coral (Corallium rubrum). The Mediterranean red coral (C. rubrum) is a branching gorgonian whose colonies could reach 50 cm height (Tsounis et al. 2010). The red coral is one of the most long-lived inhabitants of the coralligenous, in fact it can live for more than 100 years (Tsounis et al. 2010). It is a sciaphilous species that can be found at 5–800 m depth, more commonly at 30–200 m (Costantini et al. 2010).
Corallium rubrum are currently used for jewellery together with other six species (the so-called “precious corals”): Corallium secundum, Corallium regale, Corallium elatius,
Corallium konojoi, Corallium sp. nov., and Paracorallium japonicum (Tsounis et al. 2010). Despite many genetic studies focused on red coral (Abbiati et al. 1993, Costantini and Abbiati 2006; Costantini et al. 2007a, b, 2010), phylogenetic hypothesis regarding its speciation event does not exist to date. Consequently, due to its commercial importance, the presence of a great number of species belonging its family together with the fact that it represents the single species inhabiting the Mediterranean Sea, this kind of studies could be useful to individuate which are the main drivers of the C. rubrum speciation within the Mediterranean. Moreover, these information could be extremely important to analyse the evolutionary potential of this precious species in a context of global environmental change.
Two main scenarios were investigated in this thesis work. The first hypothesis supposed that the recent (after MSC, late Miocene about 5 millions years ago; Paternello et al. 2007) Mediterranean geological events promoted the allopatric speciation process of Atlantic species within the basin. The alternative scenario proposed the earlier (before MSC) and external C. rubrum birth previous the origin of the present-day Mediterranean Sea conformation, followed by secondary colonization within the basin.
1.5. Aims
The aim of this thesis work is to carry out a Mediterranean octocorals species molecular phylogenetic study using two mitochondrial genes with different mutation rate (mtMSH and 16S). Moreover, an extended datasets with Atlanto/Pacific cogeners of Mediterranean Octocorallia species were used to understand their phylogenetic relationships and to estimate the divergence times of Mediterranean species from the cogeners. Finally, the genetic relationships among Corallium rubrum and the other
Corallium species, together with a dated phylogeny were carried out to increase the knowledge about this precious Mediterranean coral. The principal outcome of the work allow to: 1) find differences and/or similarity among molecular phylogeny and morphological taxonomy of octocorals; 2) reconstruct the cladogenesis and 3) evaluate the speciation time of the Mediterranean species.
2. Materials and methods
2.1. Sampling
Molecular analyses were performed on 15 Mediterranean octocoral species (Table 2.1). These specimens were collected along the Mediterranean coasts during oceanographic campaigns (e.g. Monitoring of Calabria Marine Biodiversity; Deep red coral project) and Scuba diving activities. All the species were identified by Marzia Bo (Genova University) following the Bayer (1981) taxonomic keys. Due to difficulties related to the mtMSH region amplification, sequences of Alcyonium palmatum and
Swiftia pallida available in GenBank (http://www.ncbi.nlm.nih.gov /Entrez/) were added to the Mediterranean data set (see Table 2.1).
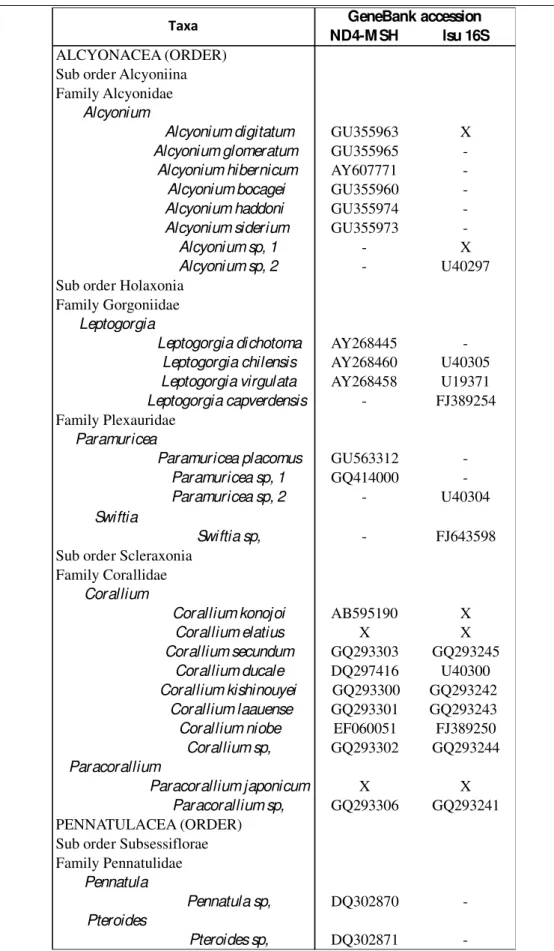
In order to evaluate the phylogenetic relationship between Mediterranean and oceanic species, Atlantic or closely related species (cogeners) sequences were added from GenBank for both markers (Table 2.2). Within the large amount of sequences published for species belonging to Alcyonium (McFadden et al. 2011) and Leptogorgia (Lepard 2003) genera, only few widespread and Atlantic distributed species were chosen for the analysis. Unspecific sequences of the analysed genera were also included to extend the sample size. Moreover, four cogeners species were analysed in laboratory. Alcyonium digitatum were obtained thanks to the Sven Lovén Centre for Marine Sciences (Göteborg University). Alcyonium sp.1 were sampled from the oceanographic vessel of I.S.P.R.A "Astrea". Corallium konojoi, Corallium elatius and
Paracorallium japonicum were obtained by Nozomu Iwasaki (Usa Marine Biological Institute, Kochi University). The combined dataset on cogeners species was generated using fewer sequences than single gene tree. The more representative analyses were performed on Corallidae family counting 8 species for both markers. For each species
just one individual were sampled. All the species were conserved in 80% ethanol at 4°C.
mtM SH 16S
ALCYONACEA (ORDER) Sub order Alcyoniina Family Alcyonidae
Alcyonium
Alcyonium palmatum GU355971 X
Alcyonium acaule X X Parerythropodium Parerythropodium coralloides X X Family Paralcyonidae Paralcyonium Paralcyonium spinulosum X X Maasella Maasella edwardsii X X
Sub order Holaxonia Family Gorgoniidae Eunicella Eunicella cavolini X X Eunicella singularis X X Eunicella verrucosa X X Leptogorgia Leptogorgia sarmentosa X X Family Plexauridae Paramuricea Paramuricea clavata X X Swiftia Swiftia pallida JQ241247 X
Sub order Scleraxonia Family Corallidae
Corallium
Corallium rubrum X X
Sub order Stolonifera Family Clavulariidae Sarcodictyon
Sarcodictyon catenatum X X
PENNATULACEA (ORDER) Sub order Subsessiflorae Family Pennatulidae Pennatula Pennatula rubra X X Pteroides Pteroides spinosum X X GeneBank accession numbers Taxa
Table 2.1. Taxonomical classification and Genbank accession numbers for the 15 Mediterranean Ottocorallia analysed. The X indicates sequences obtained in this thesis. GU355971 and JQ241247 derived from McFadden et al. (2011) and Quattrini and Cordes (2011) respectively.
ND4-M SH lsu 16S
ALCYONACEA (ORDER) Sub order Alcyoniina Family Alcyonidae
Alcyonium
Alcyonium digitatum GU355963 X
Alcyonium glomeratum GU355965
-Alcyonium hibernicum AY607771
-Alcyonium bocagei GU355960
-Alcyonium haddoni GU355974
-Alcyonium siderium GU355973
-Alcyonium sp, 1 - X
Alcyonium sp, 2 - U40297
Sub order Holaxonia Family Gorgoniidae
Leptogorgia
Leptogorgia dichotoma AY268445
-Leptogorgia chilensis AY268460 U40305
Leptogorgia virgulata AY268458 U19371
Leptogorgia capverdensis - FJ389254 Family Plexauridae
Paramuricea
Paramuricea placomus GU563312
-Paramuricea sp, 1 GQ414000
-Paramuricea sp, 2 - U40304
Swiftia
Swiftia sp, - FJ643598
Sub order Scleraxonia Family Corallidae
Corallium
Corallium konojoi AB595190 X
Corallium elatius X X
Corallium secundum GQ293303 GQ293245
Corallium ducale DQ297416 U40300
Corallium kishinouyei GQ293300 GQ293242
Corallium laauense GQ293301 GQ293243
Corallium niobe EF060051 FJ389250
Corallium sp, GQ293302 GQ293244
Paracorallium
Paracorallium japonicum X X
Paracorallium sp, GQ293306 GQ293241 PENNATULACEA (ORDER)
Sub order Subsessiflorae Family Pennatulidae Pennatula Pennatula sp, DQ302870 -Pteroides Pteroides sp, DQ302871 -GeneBank accession Taxa
Table 2.2. Genbank accession numbers of the Atlanto/Pacific species used for the analysis. The "X" indicates sequences obtained in this thesis, while "-" indicates sequences not available.
2.2. DNA extraction, amplification and sequencing
Total genomic DNA was extracted from two to four polyps per individual colonies using the cetyltrimethyl ammonium bromide protocol (CTAB), a method successfully implemented on many other marine invertebrate (e.i. Coffroth et al. 1992; Winnepenninckx et al. 1993). The CTAB protocol was adapted to laboratory contrains excluding the phenol/chloroform phase due to toxicity, without decreasing the
extraction quality. DNA was suspended in 25 µL TE buffer (Tris/EDTA solution) and conserved at -20°C until amplification. Each extraction was checked on 0.8% agarose electrophoresis gel.
Two portion of mitochondrial DNA were amplified: the mitochondrial mutS homolog gene (mtMSH, Pont-Kingdon et al. 1995) 5’ end and a portion of the
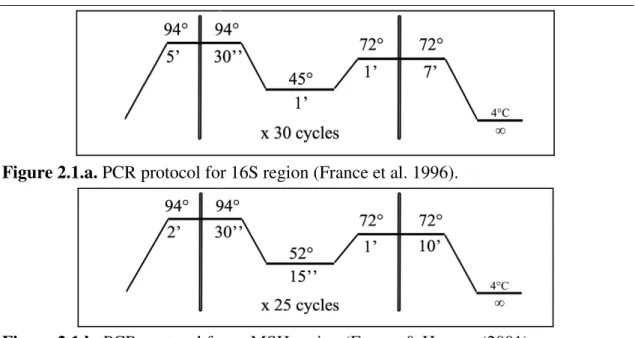
ribosomal large sub-unit (16S). mtMSH and 16S were amplified respectively using the primers ND42599F GCCATTATGGTTAACTATTAC-3') and MUT3458R (5'-TSGAGCAAAAGCCACTCC-3') designed by France and Hoover (2002); and the primers 16S Octo1L AGACCCTATCGAGCTTTACTGG-3') and 16 Octo2H (5'-CGATAAGAACTCTCCGACAATA-3') implemented by France et al. (1996). The PCR amplification was performed for 16S and mtMSH in all species applying respectively the France et al. (1996) and France and Hoover (2001) protocols
parameters (Figure 2.1.a and 2.1.b). Each PCR product was checked on 1.5% agarose electrophoresis gel. PCR products were sent to Macrogen Europe (NL) for purification and sequencing.
Figure 2.1.a. PCR protocol for 16S region (France et al. 1996).
Figure 2.1.b. PCR protocol for mtMSH region (France & Hoover (2001).
2.3. Data analyses
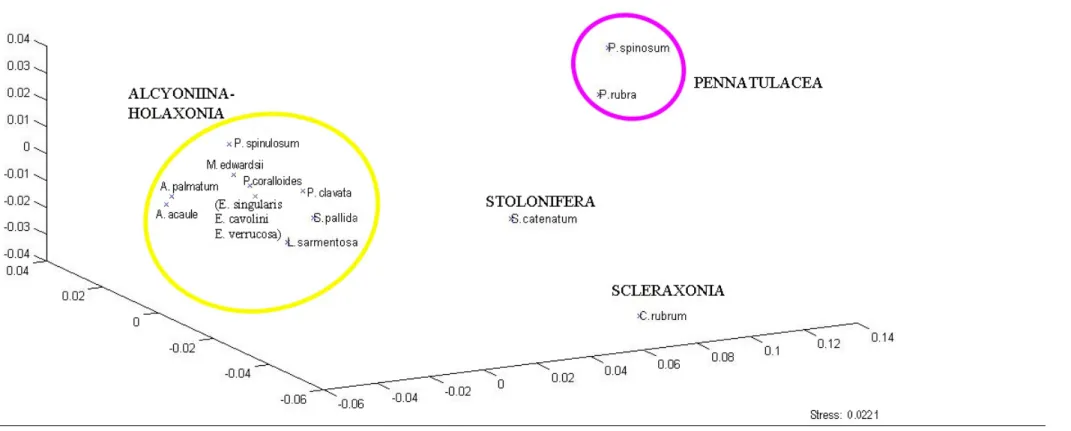
All the mtMSH and 16S sequences were edited to assess the goodness of sequencing using CROMASLITEv2.01 software (Technelysium Pty Ltd 2007). Researches of genetic similarities were performed in BLAST (Altschul et al. 1990) to identify the identity of the obtained sequences. Sequences were aligned using CLUSTALW software (Larkin et al. 2007) implemented in MEGAv.5.05 workspace (Tamura et al. 2011); the final alignment was manually performed. Number of polymorphic sites, haplotype and nucleotide diversity were obtained using DNASPv.5 (Librado et al. 2009). Distance matrixes of sequence divergence were calculated as p-distance (Dp) in MEGAv.5.05. Non-metric multi-dimensional scaling plots of distance matrixes for each marker for the Mediterranean dataset were obtained by MATLABv.7.10.0 (MathWorks 2010) and edited using PHOTOSHOP CS5.
2.4. Alignment methods
It has been repeatedly shown that the alignment quality may have an enormous impact on the final phylogenetic analysis (Tavelara and Castresana 2007). This is particularly true when comparing very divergent and unequal in length sequences, where the introduction of gaps by insertion or deletion (INDELS) in the alignments is necessary (Tavelara and Castresana 2007).
In order to find gaps and homologues sites, algorithms attempt to
approximate the INDELS reconstruction allowing lower costs for gap initiation and gap extension (Pons et al. 2006).The INDELS number decrease with higher gap opening costs (GOC), and the gap sizes increase with lower gap extension cost (GEC; Pons et al. 2006). Therefore, in both markers INDELS regions were found imposing high transition cost, small gap opening penalty and little gap extension cost in CLUSTALW algorithm (Tr cost= 1; GOC = 4, GEC= 2 in pairwise and multiple alignment; ratio GEC/GOC=0.5; implemented in MEGA 5.05).
Moreover, length variable DNA sequences such as introns and ribosomal RNA genes require alignment to establish homologous nucleotide positions for phylogenetic analysis (Pons et al. 2006; Sànchez et al. 2003a) using some highly conserved or coding sequences. Concerning the mtMSH alignment, sequences were traduced using the coelenterate mitochondrial code implemented in MEGAv.5.05. Open Reading Frames were checked by ORF-FINDER
(www.ncbi.nlm.nih.gov/projects/gorf/) adding a start codon at the beginning of each sequence. Improvement of the mtMSH alignment was achieved implementing the protein coding sequence of Sarcophyton glaucum AAC16386 (Pont-Kingdom et al. 1998) according to McFadden et al. (2005). Biases in the open reading frame were visually inspected and correct by hand in MEGAv.5.05. Backward from translation,
INDELS were localized in the mtDNA alignment using S. glaucum nucleotide
sequence AB665479 (Aratake et al. 2012), which traduces the same peptide sequence of the nucleotide sequences AAC16386. In this alignment, octocoral mtMSH
sequences identified protein functional sites (Bilewitch e Degnan 2011).
Counterpart, Ottocorallia 16S sequences were aligned using the Escherichia
coli homologous molecule (GenBank accession number: J01695, included in the operon sites 1268-2809) to identify the conserved motifs (Sànchez et al. 2003a). Likewise the mtMSH alignment, the same restricted algorithm was applied to find INDELS (Tr cost= 1; gap opening penalty = 4, gap extension cost= 2 in pairwise and multiple alignment; GEC/GOC=0.5).
The aligned nucleotide sequences of both regions were used as markers for octocoral phylogeny. The included INDELS were used as missing data in all input files.
2.5. Substitution model test and gene partition
Phylogenetic trees were developed both using each gene alignment independently and creating a concatenated dataset of both genes. Phylogenetic inference depends on the underlying evolutionary model (Guindon et al. 2009). The models that better fits the data were implemented to obtain the higher confidence in inferences (Posada et al. 2005). The best-fit substitution model selection for each dataset was calculated using JMODELTESTv.1.1 (Posada 2008) Mac software, considering 88 substitution models by hLRT calculator with 4-gamma category. The resulting models were applied to both each single gene trees and combined genes and in both trees built using the partitioned prior substitution model.
However, the combined genes maximum-likelihood analysis actually does not allow the partition of the substitution models for combined dataset, so is it requires
the single new substitution model calculated on combined sequences (Swofford 2002). The combined analysis of genes or gene-complexes (i.e., linked genes such as the mitochondrial ones) with incongruent evolutionary histories can produce misleading results by increasing the wrong phylogenetic trees support (Kubatko and Degnan 2007; Herrera et al. 2010). To quantify the conflicts that can occur between sets of characters from different data sources (e.g. the two regions mtMSH + 16S), Incongruence Length Difference test (ILD test, Farris et al. 1995) was performed. ILD test statistically compares the trees length in the parsimony context. It was computed after 1000 replicates without invariant sites estimate, and p-value < 10 % indicates that the combined dataset is not allowed (Darlu and Lecointre 2002). Moreover, the combined dataset (mtMSH + 16S) best-fit substitution model was calculated independently of the IDL results. PARTITIONFINDER (Lanfear et al. 2012) software was used to find the best partition scheme and substitutions models for the combined dataset accounting the partition data (Lanfear et al. 2012)
2.6. Maximum likelihood analyses
Maximum likelihood tree estimation (ML) consists in finding the
phylogenetic features (expressed as in relevance order: tree topology, branch lengths and parameters that describe substitution) that maximises the likelihood, i.e. the probability that the data were generated according to the selected (a priori)
evolutionary model (Guindon et al. 2009). The sites of the alignment were expected to evolve independently and under the same phylogeny (Guindon et al. 2009). The analysis produces a statistic for each tree, the maximum likelihood score (Lscore) that is the probability of that tree given the data set and the model. This score can be used
to statistically compare two different trees (i.e. two evolutionary hypotheses; Bertson 1998).
Because ML gives point-estimates of the tree (Nylander et al. 2004), non-parametric bootstrap is the most popular approach to reach the statistical support on ML tree (Guindon et al. 2009). It easily gives information about the stability of the tree topology (the branching order), and it helps in assessing whether the sequence data is adequate to validate the topology (Holmes 2003). Maximum likelihood analyses were executed on PAUPv.4.0B10 (Swofford 2002) using the heuristic search of random addition sequence to stepwise addition procedure and TBR branch-swapping with 20 random-addition-sequence replications. Node supports were calculated through 150 bootstrap replicates.
The sea pens species included in the analyses joined the same family, showed clear morphological synapomorphies (Williams 2011) and appear to be part of a dinstictive clade in many octocorals phylogenetic analyses of all anthozoans (rRNA 16S: France et al., 1996; Sanchez et al. 2003 and mtMSH: McFadden et al 2006). Consequently, Pennatulacea were used as out-group in all ML analyses and to root all ML trees.
2.7. Bayesian analyses
In phylogenetic analysis, an alternative method to searching for the single highest point in the "parameter landscape" (e.i. heuristic search in ML) is to use what is called “ marginal estimation” by Bayesian inference (Nylander 2004). Bayesian analyses make interference about the posterior probability (BP), branch lengths and other parameters, given the data, a model of character evolution and prior probabilities on parameters in the model (Nylander 2004). Metropolis-coupled Markov chain Monte
Carlo method (MCMC) is used to estimate the posterior probability of phylogenies when runs appear to have "converged'' to a stationary value (Nylander 2004). Here, Bayesian analyses were carried out using MRBAYESv.3.2 (Ronquist et al. 2011; Mac software). The MCMC analyses in this thesis run over 1*107 generations and were replicate for 2 runs. The sample frequencies parameter was set to 1000. The BP was calculated on 1*104 samples (1*107/1*103) less the first 25% of burn-in (2.5*103 samples). The obtained trees were summarized into 50% majority-rule consensus tree. Pennatulacea were used as out-group to root all Bayesian trees.
Moreover, extensive post-run analyses of MCMC for assessing convergence are seldom seen in phylogenetic literature (Nylander et al., 2008). Frequently used post-run method involves examining trace plots of the likelihood scores for trees sampled using the Markov chain method (Nylander et al., 2008). However, judging the success of a MCMC from the likelihood trace alone might lead to inaccurate and misleading results (Nylander et al., 2008). The convergence of trees parameters was evaluated by TRACERv.1.5 (Rambaut and Drummond 2007). Convergence was indicated as the ‘‘straight hairy caterpillar” (Drummond et al., 2007) shape of the stationary posterior-distribution trace (generations vs. LnL) of each parameter and it was supported from high value of the effective sample sizes ESS (>200). Other
convergence tests and mixing diagnostics were executed in the program AWTY (http:// ceb.csit.fsu.edu/awty; Nylander et al., 2008)
Finally, the phylogenetic trees obtained both from ML and Bayesian analyses were edited by FIGTREEv.1.3.1 (Rambaut 2012) and corrected by hand with
2.8. Estimation of divergence time
A Bayesian statistics method was implemented to estimate the divergence time between Mediterranean species to the other Atlanto-Pacific Ottocorallia species. An estimation of gene genealogy and divergence times was performed in BEAST 1.7.1 (Drummond et al. 2012) for the two mitochondrial marker assuming the substitution model of previous analyses.It was calculated using BEAST (Drummond and Rambaut 2007) analyses package with an uncorrelated relaxed lognormal molecular clock model was assumed. It allows for the variation in mutation rates among branches,
implementing as the tree priors the Yule model of constant speciation rate (Yule 1925; Gernhard 2008) and the coalescent model of constant population size (Kingman 1982). MCMC analyses run over 5*107 generations sampling every 5*103 before to construct 50% consensus trees by TREEANNOTATORv.1.7.1 (Rambaut and Drummond 2007).
TRACERv.1.5 (Rambaut and Drummond 2007) was used as post analyses to evaluate the convergence of trees parameters. Other examined convergence and mixing diagnostics included the standard deviation of partition frequencies (<0.01), the
potential scale reduction factor (PSRF) (ca. 1.00), the effective sample sizes (EES) (>200) and the posterior probabilities of specific nodes similitude between different runs in the program AWTY (http://ceb.csit.fsu.edu/awty) (Nylander et al. 2008).
Two ways to estimate the divergence time was used. First, the estimated mutation rates found in bibliography was used to calibrate the dated phylogeny. The 16S mutations rate was set to 0.1% substitution every million year (Palumbi 2012), while in the case of mtMSH it was set to 0.25% (Lepard 2003). Addionally, some of the few fossils available for Octocorallia, such as Coralliidae, were implemented as calibrating point. The Coralliidae node was calibrated implementing a normal prior distribution for the time to the most recent common ancestor (TMRCA) with a mean of
83.5 million years before present (Myr BP) and a standard deviation of 0.7,
corresponding to the Campanian age stratum, in which the oldest known fossil in this family has been found (Schlagintweit & Gawlick 2009). Then trees were edited by FIGTREEv.1.3.1 (Rambaut 2012) and fine-tuned with PHOTOSHOP CS5.
3. Results
3.1. Mediterranean MtMSH genetic variability
The obtained MtMSH fragments length ranged from 594 to 612 base pairs (bp hereafter) in the 15 Mediterranean species analysed. The alignment length was 627 bp including the INDELS (see the following paragraph), with 591 homologues sites excluding gaps and missing data. This alignment presented 215 polymorphic sites (36.4%), with values of haplotype diversity of 0.971 and nucleotide diversity of 0.105.
Differences in sequences length were attributable both to two large
insertion/deletion regions (INDELS) localized between the sites 90-115 and 306-316, and to two small regions (405-409 and 573-577) of the nucleotide alignment (Figure 3.1.a). Corallium rubrum and Pennatula rubra had the longest sequences (612 bp) and presented the first (90-115) and the second (306-316) big INDELS respectively. The other two smaller INDELS regions were due to the three nucleotide bases deletion in
Sarcodyction catenatum, Corallium rubrum, Pteroides spinosum and Pennatula rubra. The three Eunicella species sequences were the same length of 606 bp (Dp=0;
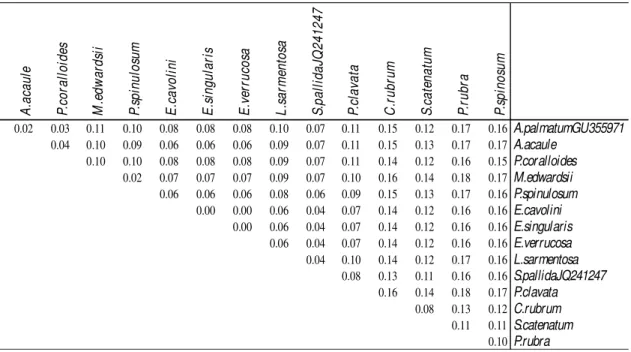
Table 3.1 and Figure 3.1.b). The main differences among sequences (as p-distance Dp>
0.15) were observed between the two order Pennatulacea and the Alcyonacea. Specifically, Paramuricea clavata and Maasella edwardsii versus Pennatula rubra presented the biggest pairwise genetic p-distances (Dp>=0.18; Table 3.1 and Figure
3.1.b). Corallium rubrum also shows great genetic distance compared to all the other species ranging between Dp=0.13 and Dp=0.16, particularly against P. clavata and M. edwardsii (Dp=0.16; Figure 3.1.b).
[ 111111111122222222223333333333444444444455555555556] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e CAAATTGGTAAGTTCTATGAACTTTGGCACGAGCCCGATACTTCTAGTAGGCAACAAGCA A. pal mat umGU355971 . . . . P. cor al l oi des . . G. . . T. . . A. . . . M. edwar dsi i . . G. . . T. . . T. . . C. . . . C. . . . P. spi nul osum . . G. . . G. . . T. . . T. . . C. . . . E. cavol i ni . . . T. . . TA. . . C. . . . E. si ngul ar i s . . . T. . . TA. . . C. . . . E. ver r ucosa . . . T. . . TA. . . C. . . . L. sar ment osa . . G. . . T. . . T. . . CG. . . . S. pal l i daJQ241247 . . G. . . T. . . T. . . . P. cl avat a . . G. . . T. . . T. CC. . . C. . . . C. r ubr um . . G. . . TA. . . . G. . . GT. C. . . GTA. . . . S. cat enat um . . G. . . T. . . G. . . T. . . GT. . . G. . . A. . . . P. r ubr a . . G. . . T. . . T. . . G. . C. . T. . . TA. . G. . . . P. spi nosum . . G. . . T. . . T. . . GT. C. . T. . . TA. . G. . . . [ 111111111111111111111] [ 666666666777777777788888888889999999999000000000011111111112] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e TACTCTCAAGCTGAGCTATTAGCTGAATCATCCATG- - - CGAAGTCAGCCT A. pal mat umGU355971 . . . - - - . . . . P. cor al l oi des . . . T. . . - - - . . . C. . . . M. edwar dsi i . . . . T. . . C. . . T. . . T. . . G. . . - - - . . . GG. T. . P. spi nul osum . . . . T. . . C. . . T. . . T. . . G. . . - - - . . . GG. T. . E. cavol i ni . . . C. . . T. . . T. . . G. . . - - - . . . GG. . . . E. si ngul ar i s . . . C. . . T. . . T. . . G. . . - - - . . . GG. . . . E. ver r ucosa . . . C. . . T. . . T. . . G. . . - - - . . . GG. . . . L. sar ment osa C. . A. . . . G. . C. . . T. . . T. . . GC. . . - - - . . . AG. . . . S. pal l i daJQ241247 . . . C. . . T. . . T. . . G. . . - - - . . . G. . . . P. cl avat a . . . C. . . T. . . T. . . G. . . - - - . . . GG. . . . C. r ubr um . . . T. . . T. . . G. . GC. T. . ACCAATTTACATA. . . AG. . . . S. cat enat um . . . T. . G. . . T. A. G. . G. . . A- - - . . . GG. . . . P. r ubr a C. T. . . T. . . T. . TG. . GGG. CCCCCCGTG- - - . . GGG. . . . P. spi nosum . . T. A. . . T. . . A. . . G- - - GCCCCT- - - GG. . . . [ 111111111111111111111111111111111111111111111111111111111111] [ 222222222333333333344444444445555555555666666666677777777778] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e TTGGGGGTAACGCCCCCCATTGAACAAGTTGCCTCATTACTTGATATGAGAATAATATTG A. pal mat umGU355971 . . . . P. cor al l oi des . . . . M. edwar dsi i . . . G. . . A. . T. . . G. . . CG. C. P. spi nul osum . . . G. . . A. . T. . . G. . . CG. C. E. cavol i ni . . . . A. . . T. . . G. . . C. E. si ngul ar i s . . . . A. . . T. . . G. . . C. E. ver r ucosa . . . . A. . . T. . . G. . . C. L. sar ment osa . . . . A. . . A. . . A. . . C. S. pal l i daJQ241247 . . . T. . . A. . . G. . . C. P. cl avat a . . . . A. . . T. . . A. . . C. . CA C. r ubr um . . . A. . T. . . G. . . G. . . C. . . . A. . . CA S. cat enat um . . . A. . . G. . . CA P. r ubr a . . A. AAA. . . . T. . T. . T. . . T. . . G. . . CC P. spi nosum . . A. . . A. . A. . . T. . . G. . . G. . . CA
[ 111111111111111111122222222222222222222222222222222222222222] [ 888888888999999999900000000001111111111222222222233333333334] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e CCCGGTAAAAGATCTTTGCTTCAAATGGGATTTCCAATTTATTCCCTTACTACTCATCTA A. pal mat umGU355971 . . . . P. cor al l oi des . . . G. . . T. . M. edwar dsi i . . . C. . . G. . . G. C. . . . P. spi nul osum . . . C. . . G. . . G. C. . . . E. cavol i ni . . . CC. . . G. . . . E. si ngul ar i s . . . CC. . . G. . . . E. ver r ucosa . . . CC. . . G. . . . L. sar ment osa . . . C. . . C. . . G. . . . S. pal l i daJQ241247 . . . C. . . . A. . . C. . . G. . . . P. cl avat a . . . CC. . . G. . . CG. . . G. C. r ubr um . . . C. . . G. . . T. . A. . . T. . . A. . S. cat enat um . . . C. . . CC. . . G. . . TG. . . T. . P. r ubr a . . . T. . . G. . . T. . . T. . CG. . . T. . T. . P. spi nosum . . T. . . C. . . G. . . T. C. . . AC. . . T. . T. . [ 222222222222222222222222222222222222222222222222222222222223] [ 444444444555555555566666666667777777777888888888899999999990] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e AGCACCCTATTGGATAAAGGTTGGACTGTTATAGTTATCGATGAATTAGTCACTGGTAAA A. pal mat umGU355971 . . . . P. cor al l oi des . . . T. . . A. . . . M. edwar dsi i . . T. . TT. G. . . . P. spi nul osum . . T. . TT. G. . . AA. . . . E. cavol i ni . . T. . . T. G. . . T. . . T. . . . E. si ngul ar i s . . T. . . T. G. . . T. . . T. . . . E. ver r ucosa . . T. . . T. G. . . T. . . T. . . . L. sar ment osa . . T. . TT. G. . . T. . . C. . . S. pal l i daJQ241247 . . T. . TT. G. . . T. . . . P. cl avat a . . T. . . T. GC. . . T. . . GA. . . . C. r ubr um . . T. . T. . G. . . G. . . TA. . . A. . . C. . T. . . . S. cat enat um . . . TT. G. . . A. C. . . T. . . T. . . . P. r ubr a . . . CT. T. G. . . A. . . T. . . C. . . . T. . . . P. spi nosum G. . . . TT. G. . . A. . . C. . T. . . C. . . C. . . [ 333333333333333333333333333333333333333333333333333333333333] [ 000000000111111111122222222223333333333444444444455555555556] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e TCCGGG- - - CCAAAACAACGTGCAGTATCTCAGGTTTATTCTCCTAGTTGTAAT A. pal mat umGU355971 . . . - - - . . . . P. cor al l oi des . . . - - - . . . . M. edwar dsi i . . A. . . - - - . . . C. . . . P. spi nul osum . . A. . . - - - . . . C. . . . E. cavol i ni . . T. . . - - - . . . C. . . C. . . . E. si ngul ar i s . . T. . . - - - . . . C. . . C. . . . E. ver r ucosa . . T. . . - - - . . . C. . . C. . . . L. sar ment osa . . T. . . - - - . . . C. . . . S. pal l i daJQ241247 . . T. . . - - - . . . C. . . . P. cl avat a . . . - - - . . . C. . . . C. r ubr um . . G. . . - - - . . G. . . C. . . G. . . . C. . . S. cat enat um . . A. . . - - - . . C. . . C. . . . P. r ubr a . . . . A. CCCAAACAG. . C. . . C. . . G. . . . AA. . . T. . . . P. spi nosum . . . A- - - . . T. . . C. . . C
[ 555555555555555555555555555555555555555555555555555555555556] [ 444444444555555555566666666667777777777888888888899999999990] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e TGGGCCAACTCAGGGGTTGGCTCAGATATTTTAATAAATAAAATATATAATTTATTAATT A. pal mat umGU355971 . . . G. . A. CC. . . G. . . . P. cor al l oi des . . . G. . A. CC. A. . . G. . . . M. edwar dsi i . . . . TA. . . . TG. . . C. . . G. C. . . G. . . C. . . . P. spi nul osum . . . . TA. . . . TG. . . C. . . . G. . . G. . . . E. cavol i ni . . . . TA. . . . E. si ngul ar i s . . . . TA. . . . E. ver r ucosa . . . . TA. . . . L. sar ment osa . . . . TAG. . . G. . . G. . . . S. pal l i daJQ241247 . . . . GAG. . . G. . . . P. cl avat a . . . A. . . G. . . A. . . G. . . GG. . . C. . . . C. r ubr um . . . GG. . CTG. . . . CCAAT. . . . GA. . AC. . - - - . . . G. . . GG. . . . S. cat enat um . . . GG. . . . G. . . . CC. AT. . . . GG. . AC. . - - - . . . G. . . GG. . . . P. r ubr a . . . GG. . . . G. . . . CC. ATC. . . GG. . AC. . - - - . . . G. . . GG. . . . P. spi nosum . . . . TGG. . . . G. . . . CC. ATC. . . GA. . A. . . - - - . . . G. . . GG. . . . [ 666666666666666666666666666]
[ 000000000111111111122222222] [ 123456789012345678901234567] A. acaul e GGTTGGAATTTATTCCCCCCTGAACCC A. pal mat umGU355971 . . . T. . . . P. cor al l oi des . . . T. . . T. . . . M. edwar dsi i . A. . . C. . . . P. spi nul osum . A. . . . E. cavol i ni . . . . E. si ngul ar i s . . . . E. ver r ucosa . . . . L. sar ment osa . . . A. . . T. . . . G. . . S. pal l i daJQ241247 . . . T. . . . G. . . P. cl avat a . . . CC. . . G. . . C. r ubr um . . C. . . TT. . . . G. . T S. cat enat um . . . T. . . . G. . T P. r ubr a . . C. . . G. . . TT. . . T P. spi nosum . . . T. . . T
Figure 3.1.a. Alignment of MtMSH sequences of the 15 Mediterranean species analysed. Points mean identical bases to those of Alcyonium acaule sequence; dashes mean deletions. A .a ca u le P. co ra ll o id es M .e d w a rd si i P. sp in u lo su m E .c a vo li n i E .s in g u la ri s E .v er ru co sa L .s a rm en to sa S .p a ll id a JQ 2 4 1 2 4 7 P. cl a va ta C .r u b ru m S .c a te n a tu m P. ru b ra P. sp in o su m 0.02 0.03 0.11 0.10 0.08 0.08 0.08 0.10 0.07 0.11 0.15 0.12 0.17 0.16A.palmatumGU355971 0.04 0.10 0.09 0.06 0.06 0.06 0.09 0.07 0.11 0.15 0.13 0.17 0.17A.acaule 0.10 0.10 0.08 0.08 0.08 0.09 0.07 0.11 0.14 0.12 0.16 0.15P.coralloides 0.02 0.07 0.07 0.07 0.09 0.07 0.10 0.16 0.14 0.18 0.17M.edwardsii 0.06 0.06 0.06 0.08 0.06 0.09 0.15 0.13 0.17 0.16P.spinulosum 0.00 0.00 0.06 0.04 0.07 0.14 0.12 0.16 0.16E.cavolini 0.00 0.06 0.04 0.07 0.14 0.12 0.16 0.16E.singularis 0.06 0.04 0.07 0.14 0.12 0.16 0.16E.verrucosa 0.04 0.10 0.14 0.12 0.17 0.16L.sarmentosa 0.08 0.13 0.11 0.16 0.16S.pallidaJQ241247 0.16 0.14 0.18 0.17P.clavata 0.08 0.13 0.12C.rubrum 0.11 0.11S.catenatum 0.10P.rubra
Table 3.1. Pairwise genetic p-distance (Dp) among Mediterranean species of MtMSH sequences.
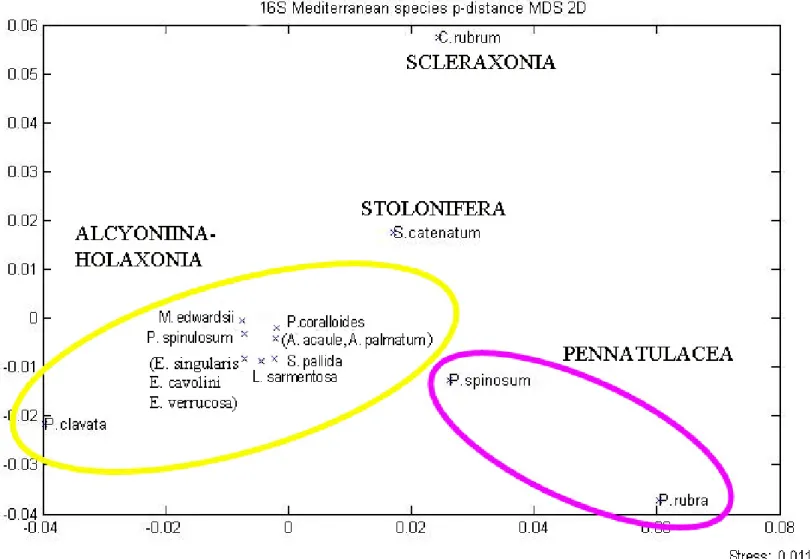
3.2. Mediterranean 16S genetic variability
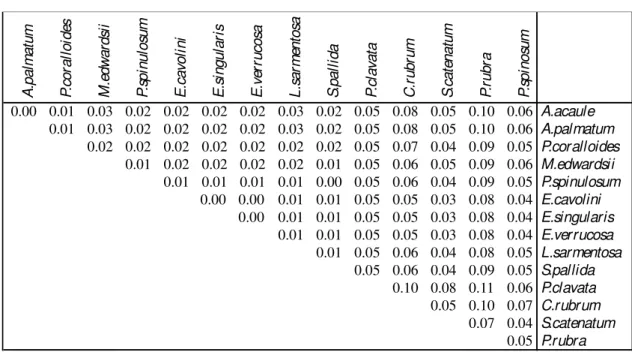
The 16S fragments length varied from 224 to 267 bp in the 15 Mediterranean taxa analysed. The length of the aligned sequences was 274 bp including the INDELS (see the following pragraph), exhibiting 219 homologues sites excluding gaps and missing data. Out of 274 pb only 37 were polymorphic sites (16.9%). The alignment presented a value of haplotype diversity of 0.952, related to a nucleotide diversity of 0.041. One single large insertions region in S. catenatum and C. rubrum represented the great source of variation between sequences length. It was localized between the homologues sites 148-201 (Figure 3.2.a).
Alcyonium acaule and A. palmatum exhibited no sequences divergence in the case 16S marker (Dp=0; Table 3.2 and Figure 3.2.b). The three sequences of the Eunicella species showed the identical nucleotide sequences (Dp=0; Table 3.2 and
Figure 3.2.b). The greatest divergence among sequences (Dp>0.11) was observed
between Pennatula rubra and Paramuricea clavata (Table 3.2 and Figure 3.2.b). As seen in MtMSH, Corallium rubrum showed higher genetic distance compared to all the other species, ranging between Dp=0.05 and Dp=0.10, particularly against P. clavata
[ 111111111122222222223333333333444444444455555555556] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e CTAATTTTGAGTTAATAATTTTT- GTTGGTGGGACAGTTTAGTTGGGGCGACTACCTTTG A. pal mat um . . . - . . . . P. cor al l oi des . . . - . . . A. . . . M. edwar dsi i . . G. . . - . . . A. . . . P. spi nul osum . . G. . . - . . . A. . . . E. cavol i ni . . . T. . . A. . . . E. si ngul ar i s . . . T. . . A. . . . E. ver r ucosa . . . T. . . A. . . . L. sar ment osa . . . - . . . A. . . . S. pal l i da . . G. . . - . . . A. . . . P. cl avat a . . . CA. . . AA. . . - . . . A. . . . C. r ubr um . . . A. . . - . . . CA. . . C. . S. cat enat um . . . C. . . - . . . A. . . . P. r ubr a . . . C. A. . . A. . . . - . . . A. . . . P. spi nosum . . . C. A. . . A. . . . - . . . A. . . . [ 111111111111111111111] [ 666666666777777777788888888889999999999000000000011111111112] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e AATAAGAAACGAAGGCGAGCTTATGGTATACAA- AGCTAATCACATTAGCCTGACAGTGA A. pal mat um . . . - . . . . P. cor al l oi des . . . - . . . . M. edwar dsi i . . . - . . . T. . . . P. spi nul osum . . . - . . . T. . . . E. cavol i ni . . . T. . - . . . T. . . . E. si ngul ar i s . . . T. . - . . . T. . . . E. ver r ucosa . . . T. . - . . . T. . . . L. sar ment osa . . . - . . . T. . . . S. pal l i da . . . - . . . T. . . . P. cl avat a . . . - . . . TT. T. . . . C. r ubr um . . . G. . . - T. TT. A- - . . . . T. . . . S. cat enat um . . . - T. TT. A- - . . . . T. . . C. . . . P. r ubr a . . . - TG. T. A- - . . . . T. . . C. . . . P. spi nosum . . . - T. . T. A- - . . . . T. T. . . . [ 111111111111111111111111111111111111111111111111111111111111] [ 222222222333333333344444444445555555555666666666677777777778] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e GGGGGACACCCCTAG CTGGCACAAGGA CGAC G A. pal mat um . . . . . . . . . . . P. cor al l oi des . . . . . . . A. . . M. edwar dsi i . . . . . . . T. . . P. spi nul osum . . . . . . . T. . . E. cavol i ni . . . . . . . T. . . E. si ngul ar i s . . . . . . . T. . . E. ver r ucosa . . . . . . . T. . . L. sar ment osa . . . . . . . T. T . S. pal l i da . . . . . . . T. . . P. cl avat a . . . . . . . T. G . -C. r ubr um . . . - . . . - - CTT. . CCAT- T- ACA- - - ACGTA S. cat enat um . . . - . . . CTT. C. . - - ATAGTACTTAGACATAGTACGTA P. r ubr a . . . . . . . CGAG. . . A. . . G CC. CG. . AG P. spi nosum . . . . . . CC. CG. . GG
-[ 111111111111111111122222222222222222222222222222222222222222] [ 888888888999999999900000000001111111111222222222233333333334] [ 123456789012345678901234567890123456789012345678901234567890] A. acaul e - - - TCCTATGTGGGCGATAATGACCCGATATGATTGTCCAAAA A. pal mat um - - - . . . . P. cor al l oi des - - - . . . . M. edwar dsi i - - - . . . A. . . . P. spi nul osum - - - . . . . E. cavol i ni - - - . . . . E. si ngul ar i s - - - . . . . E. ver r ucosa - - - . . . . L. sar ment osa - - - . . . A. . . . S. pal l i da - - - . . . . P. cl avat a - - - . T. . . A. . . . C. r ubr um - - - ATGG- - - GT. . . A. . . A. . . T. . . S. cat enat um GTGCGTAGCACGTAGTGGAT. . . . P. r ubr a - - - GGT. . . G. . . T. . . A. . . . P. spi nosum - - - G- T. . . G. . . A. . . . [ 2222222222222222222222222222222222] [ 4444444445555555555666666666677777] [ 1234567890123456789012345678901234] A. acaul e TAAATATCGAAAGCGAATAAAAGCTACCGTAGGG A. pal mat um . . . . P. cor al l oi des . . . . M. edwar dsi i . . . T. . . . P. spi nul osum . . . G. . . ?. . . E. cavol i ni . . . G. . . . E. si ngul ar i s . . . G. . . . E. ver r ucosa . . . G. . . . L. sar ment osa . . . G. . . . S. pal l i da . . . G. . . . P. cl avat a . . . . C. r ubr um . . . G. . . C. . . . S. cat enat um . . . G. . . . P. r ubr a G. . . G. . . . P. spi nosum . . . G. . . .
Figure 3.2.a. Alignment of the portion of the 16SrDNA gene for the 15 species analysed. A .p a lm a tu m P .c o ra ll o id es M .e d w a rd si i P .s p in u lo su m E .c a vo li n i E .s in g u la ri s E .v er ru co sa L .s a rm en to sa S .p a ll id a P .c la va ta C .r u b ru m S .c a te n a tu m P .r u b ra P .s p in o su m 0.00 0.01 0.03 0.02 0.02 0.02 0.02 0.03 0.02 0.05 0.08 0.05 0.10 0.06 A.acaule 0.01 0.03 0.02 0.02 0.02 0.02 0.03 0.02 0.05 0.08 0.05 0.10 0.06 A.palmatum 0.02 0.02 0.02 0.02 0.02 0.02 0.02 0.05 0.07 0.04 0.09 0.05 P.coralloides 0.01 0.02 0.02 0.02 0.02 0.01 0.05 0.06 0.05 0.09 0.06 M.edwardsii 0.01 0.01 0.01 0.01 0.00 0.05 0.06 0.04 0.09 0.05 P.spinulosum 0.00 0.00 0.01 0.01 0.05 0.05 0.03 0.08 0.04 E.cavolini 0.00 0.01 0.01 0.05 0.05 0.03 0.08 0.04 E.singularis 0.01 0.01 0.05 0.05 0.03 0.08 0.04 E.verrucosa 0.01 0.05 0.06 0.04 0.08 0.05 L.sarmentosa 0.05 0.06 0.04 0.09 0.05 S.pallida 0.10 0.08 0.11 0.06 P.clavata 0.05 0.10 0.07 C.rubrum 0.07 0.04 S.catenatum 0.05 P.rubra
3.3. Mediterranean phylogenetic analysis
3.3.1 MtMSH Mediterranean trees
The General Time Reversible model (GTR) with gamma distribution (+G) more invariant sites (+I) resulted to be the MtMSH dataset best substitution model. The shape of the gamma distribution was set to 0.667 and the proportion of invariant sites was 0.116. TRACER results on Bayesian tree show high value of ESS (>200) and AWTY post analyses highly supported the convergence of tree parameters.
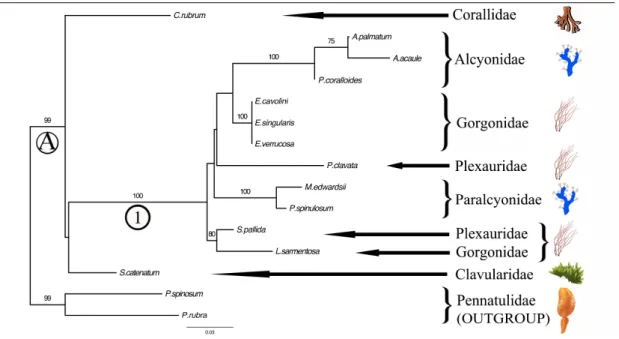
Both Maximum-likelihood and Bayesian analyses recovered similar trees topologies (Figure 3.3.a and 3.3.b respectively). The out-group clade comprised the two sea pens as a monophyletic group (BT=99% and BP=100%; Figures 3.3). The main cluster (clade A; Figures 3.3) containing all the analysed species excepted the sea pens (out-group), was well supported by the maximum-likelihood bootstrap value (BT=99%) and Bayesian posterior probabilities (BP=100%).
At sub-order level within the Clade A, a large sub-clade (clade 1) included all members of the sea fan order Holaxonia and the soft corals belonging to sub-ordinal group Alcyoniina, both linked as paraphyletic lineages (BT=100% and BP=100%; Figures 3.3). Moreover, analysed Stolonifera (S. catenatum) and Scleraxonia (C. rubrum) were linked at the base of the Clade A without clear relationships with the Clade 1.
At lower taxonomic level (family), the Clade 1 showed three monophyletic groups: Paralcyonidae, Alcyonidae and Gorgonidae family. The Paralcyonidae family appeared to be an independent lineage (BT=100% and BP=100%) and did not join with the other soft coral belonging to the Alcyonidae family. The Mediterranean Alcyoniadae species belonging to the two analysed genera Alcyonium and