56
CHAPTER IV
57 1. Sample purification from endogenous DNA contamination
For testing the performances of sample filtration process, the presence of residual endogenous swine gene was evaluated by PCRs: a less intense DNA fragment of expected size was detected in filtered serum sample than in no filtered sample, demonstrating that filtration process reduced the presence of host contaminant DNA (Fig. 8A).
Subsequently, the same PCRs were performed to evaluate the filtration followed by Benzonase –treatment on pig serum, using 50U of enzyme. No DNA fragments were observed on agarose gel, demonstrating that the treatment with Benzonase seemed to be necessary to completely remove any host DNA contamination from the sample (Fig. 8B). For demonstrating that the filtration process followed by treatment with enzyme did not affect the presence of TTSuV genome in the sample, a common PCR to detect TTSuV2 DNA was done (Fig. 9).
8A) 8B)
Fig. 8 Endogenous swine gene DNA presence in serum sample TTSuV1 and TTSuV2 positive. A) PCR carried out with Glucagon gene primers (GLC-F and GLC-R) on no filtered (lane 1) and filtered (lane 2) serum. Symbol “–” represents the negative control (sterile-filtered water); symbol “+” represents the positive control (serum
58 positive for the presence of endogenous swine gene, no filtered). B) PCR carried out with Phospholipase C (pLCb-1-F and pLCb-1-R) and βLactoglobulin gene (βLG-F and βLG-R) primers. M1 = 100 bp DNA molecular marker.
Fig. 9 TTSuV2 DNA presence in serum no filtered/no Benzonase-treated (lane 1) and in serum filtered and treated (lane 2). Symbol “–” represents the negative control (sterile-filtered water); symbol “+” represents the positive control (pig serum positive for TTSuV1 and TTSuV2).
2. RCA-SISPA in swine
The combined RCA-SISPA method resulted in several fragments of various lengths: fragments between 300 bp and 3 Kb were isolated from the agarose gel (Fig. 10). The corresponding amplified products obtained from PCR using the ligations DNA-adaptors as templates are shown in Figure 11 (A, B, C, D). After sequencing, 3 out of 66 (4.5%) fragments corresponded to TTSuV2. Other sequences included bacterial genomes (Eubacterium, Diaphorobacter and Mycoplasma), fungi genome (Sclerotinia sclerotiorum), pig genome, human DNA and cloning vector.
59 Fig. 10 Digestion products obtained by using RCA-SISPA approach. RCA products digested by TaqI and Csp6I were run on 1.3% agarose gel. M1 = 100 bp DNA molecular marker; M2 = 1Kb DNA molecular marker.
60 11A) 11B)
11C) 11D)
Fig. 11 PCR products obtained by using ligations DNA-adaptors as templates and NTaq24 (A, B) or NBam24 (C, D) as corresponding primers. Symbol “–” represents the negative control (sterile-filtered water). Black arrows indicate the expected amplified fragments. Aspecific fragments also are observed (D). M1 = 100 bp DNA molecular marker; M2 = 1Kb DNA molecular marker.
61 3. Anello-RCA/PCR in swine and human
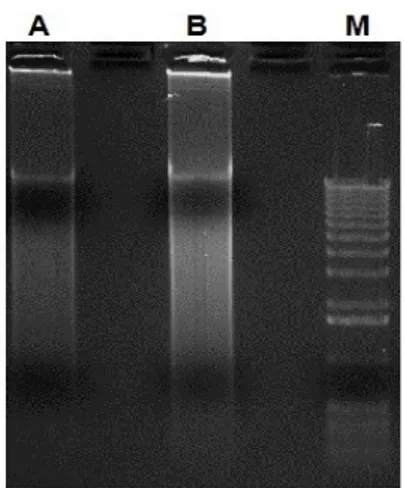
The amplification products resulting from Anello-RCA corresponded to DNA of high molecular weight, observed as a smear on agarose gel (Fig. 12).
Following Anello-PCR optimization, best conditions for swine samples were: Tm 54.5°C, 1.5 mM MgCl2 and 10-100 ng of input Anello-RCA product. With these
conditions, DNA fragments of various lengths were obtained testing swine serum previously found positive for TTSuV (Fig. 13A). Fragments of 2.7-2.9 Kb, corresponding to the expected length of TTSuV genome, were observed and then cloned for further sequence analysis.
Similarly, Anello-PCR was optimized for human TTV detection. The optimal conditions were: both Tm 40°C and 50°C, the use of La Taq Takara (2.5 U) with its Buffer II and 20 ng of Anello-RCA product. With these conditions, DNA fragments between 3 and 4 Kb in length were amplified from human serum previously found positive for TTV by “universal” quantitative PCR (Fig. 13B). To confirm the effectiveness of the Anello-PCR in human, fragments 3 and 4 Kb in size were also obtained from other 4 sera positive for TTV by quantitative PCR (Fig. 13C). Sequencing of the cloned amplified samples revealed that all of them contained TTSuVs (in pig serum) or TTVs (in human sera). Therefore, full-length genomes of TTSuV1, TTSuV2 and TTV were successfully obtained using Anello-RCA in combination with Anello-PCR.
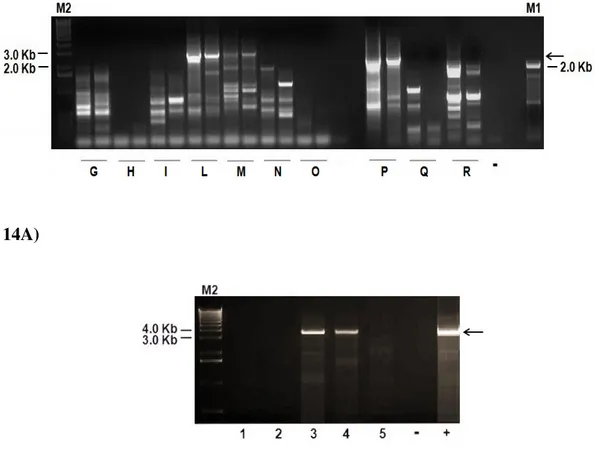
The optimized conditions were subsequently applied to serum samples previously tested negative for swine and human TTVs (Fig. 14A, 14B, respectively). From 10 PCR negative swine sera, 3 TTSuVs (one TTSuV1 and two TTSuV2) could be
62 identified. The shorter not expected fragments from swine sera observed on agarose gel (Fig. 14A) represented bacterial and host DNA sequences. In addition, 2 out of 5 human sera resulted TTV positive (tth14, No. AJ620228, 97% similarity; TTVyon-LC011, No. AB038622, 90% similarity).
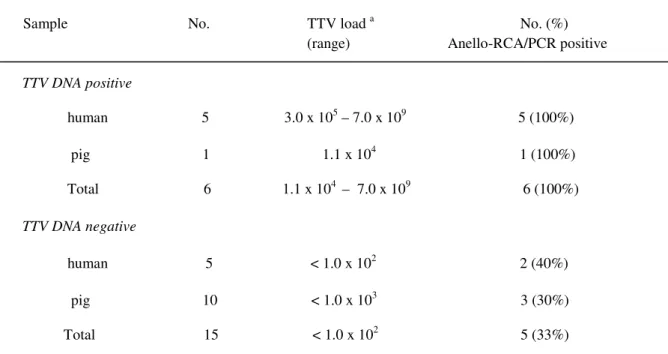
Results of testing 21 serum samples from humans and pigs for TTV genome full-length detection by Anello-RCA/PCR are summarized in table 4.
Fig. 12 Anello-RCA products (2 µl) run on 0.6% agarose gel before (lane A) and after (lane B) purification and concentration. M = 1 Kb DNA molecular marker.
63 13A) 13B)
13C)
Fig. 13 Anello-PCR products from serum samples positive for TTSuV. A) 2.7 Kb fragment corresponding to the complete TTSuV genome obtained by using the optimized Anello-PCR conditions. Different dilutions (1= 1:10, 2=1:100, 3=1:1000, 4=1:10000) of the Anello-RCA product used as template were tested. A black arrow indicates TTSuV full-length genome. The shorter fragments represent bacterial sequences. B) Anello-PCR products obtained from human TTV positive control
64 serum. Fragments between 3 and 4 Kb represent the complete genome of human TTV (lane A, from Anello-PCR with Tm 40°C; lane B, from Anello-PCR with Tm 50°C). A black arrow indicates TTV full-length genome. C) Anello-PCR products obtained from 4 human TTV positive sera samples (C, D, E, F). A black arrow indicates TTV full-length genome. Symbol “–” represents the negative control (sterile-filtered water). M1 = 100 bp DNA molecular marker; M2 = 1 Kb DNA molecular marker.
14A)
14B)
Fig. 14 Anello-PCR products from swine (A) and human (B) sera initially found negative for TTSuV and TTV, respectively. A) Anello-PCR products obtained from
65 10 TTSuV negative swine sera (lanes G, H, I, L, M, N, O, P, Q, R). Duplicates of each sample are given. B) Anello-PCR products obtained from 5 TTV negative human sera (lanes 1, 2, 3, 4, 5). The symbols “–” and “+” represent the negative (sterile-filtered water) and positive (human TTV positive serum) controls, respectively. Black arrows indicate fragments of 2.8 Kb (A) and 3.8 Kb (B) corresponding to TTSuV and TTV full-length genomes, respectively. The shorter fragments represent bacterial and host DNA sequences (A). M1 = 100 bp DNA molecular marker; M2 = 1 Kb DNA molecular marker.
Sample No. TTV load a No. (%) (range) Anello-RCA/PCR positive
TTV DNA positive human 5 3.0 x 105 – 7.0 x 109 5 (100%) pig 1 1.1 x 104 1 (100%) Total 6 1.1 x 104 – 7.0 x 109 6 (100%) TTV DNA negative human 5 < 1.0 x 102 2 (40%) pig 10 < 1.0 x 103 3 (30%) Total 15 < 1.0 x 102 5 (33%) a
TTV DNA copies per mL of serum as determined by quantitative real-time PCR.
Table 4. TTV full-length genome detection by Anello-RCA/PCR in 21 sera from humans and pigs, known to be TTV DNA positive or negative by quantitative real-time PCR.
66 4. Anello-RCA/PCR in other animal species
The amplification pattern resulting from Anello-RCA corresponded to DNAs of high molecular weight, as observed in swine and human (Fig. 12).
Tm 54.5°C, 1.5 mM MgCl2 and 10-100 ng of input Anello-RCA product were the
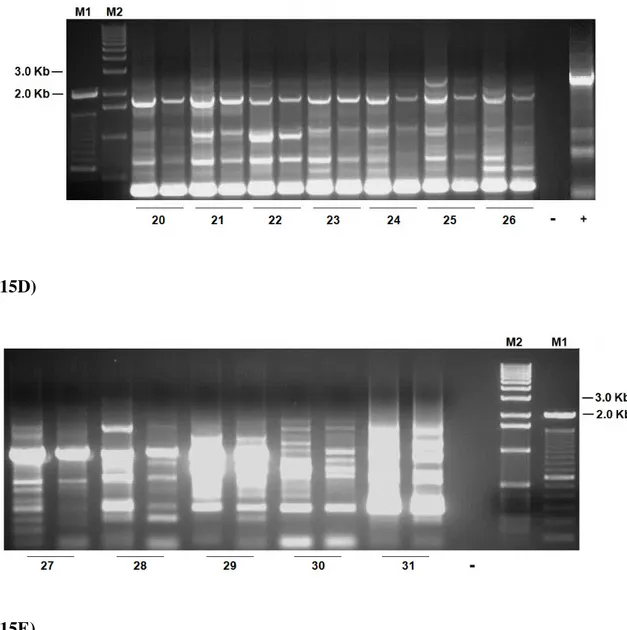
reaction conditions used in Anello-PCR previously optimized for swine serum. With these conditions, DNA fragments of various lengths were obtained from serum samples of other animal species (Fig. 15). Each fragment, different in length from the others, was cloned for further sequence analysis. No sequences related to Anelloviridae genomes were detected.
67 15B)
68 15D)
15E)
Fig. 15 Anello-PCR products from 159 serum samples from chicken, bovine, sheep and birds. Reaction conditions optimized for swine were used. The symbols “–” and “+” represent the negative (sterile-filtered water) and positive (pig TTSuV positive serum) controls, respectively. All fragments observed represent bacterial or host DNA sequences. M1 = 100 bp DNA molecular marker; M2 = 1 Kb DNA molecular marker.
69 Similar results were obtained testing 36 out of 159 serum samples by Anello-PCR optimized with conditions previously used for human TTV amplification (data not shown). Results of testing 159 sera of several animal species for TTV DNA detection by Anello-RCA/PCR are shown in table 5.
Animal species No. examined Result by Anello-RCA/PCR Birds
Pavus cristatus 9 Negative Laurus audonii 6 Negative Phoenicopterus ruber 6 Negative
Columba livia 7 Negative Anas platyrinchos 8 Negative Bovine 68 Negative Chicken 49 Negative Sheep 6 Negative Total 159 Negative
Table 5. Results of testing 159 sera from several animal species for TTV DNA by Anello-RCA/PCR.
70 5. Evaluation of Anello-RCA and RCA efficiencies by quantitative PCR
Quantitative PCRs for TTSuV1 and TTSuV2 were used to evaluate the efficiencies of Anello-RCA and RCA techniques. DNA extracted from TTSuVs positive pig serum, was tested for TTSuV2 load: a viremia level of 1.1x104 copies/mL was found. After amplification using the Anello-RCA technique, such load increased to 4.0 x109 copies TTSuV2/mL of input DNA. Subsequently, efficiencies of Anello-RCA and Anello-RCA were compared by performing simultaneous amplification of three different TTSuVs negative pig sera resulted positive by Anello-RCA (Fig. 14A). Anello-RCA from the pig serum initially negative for TTSuV1 revealed 9.1x107 copies of TTSuV1/mL while the corresponding RCA product showed 2.8x107 copies of TTSuV1/mL. Quantitative PCR for other two pig sera initially negative for TTSuV2 but positive by Anello-RCA (Fig. 14A), contained 7.1x103 copies/mL and 5.7x104 copies/mL of TTSuV2 after Anello-RCA and 4.7x103 copies/mL and 5.9x103 copies/mL of TTSuV2 after RCA.