97
4. RESULTS AND DISCUSSION
4.1 Cell growth inside the bioreactor
HMEC-1 cells have been cultured in MCDB131 medium (SIGMA) and subjected to different growing tests inside the bioreactors. For example, the European Space Agency specifically asked for pre-incubation tests to be performed at suboptimal temperature, in order to simulate possible pre-mission scenarios.
To comply with this request, HMEC-1 were incubated at 18°C or 20°C for up to 7 days, placing 37°C-heated “ice bricks” along with the experimental units (EUs) inside a polystyrene box.
The purpose of this section is to present and illustrates the results of the above mentioned tests.
All the growing tests results are listed in the table below (Tab.2), which also contains all the experimental parameters of each test: the type of culture support used, the initial number of cells seeded on it, the presence of HEPES buffer in culture medium or of the gelation coating on the coverslip and the incubation protocol. The results are expressed in terms of cell number as well.
The present data will then be discussed, focusing on the following issues one by one: cell culture support, coverslip coating, initial cell number and incubation protocol.
98 Exp # Coverslip Initial cell number (cells/cm2) Hepes 20 mM Gelatin coating (1) Protocol of Incubation (2) Results (cell number) 1 Thermanox Nunc 174934 5000 (2100 cells/cm2) No Yes 3dd@37°C + 9dd@37°C 450000 (3) 2 500 (200 cells/cm2) No Yes 7dd@16°T< 18°C Empty coverslip 3 2500 (1100 cells/cm2) No Yes 7dd@16°C T<18°C Empty coverslip 4 500 (200
cells/cm2) No Yes 7dd@18°C Empty coverslip
5 2500 (1100
cells/cm2) No Yes 7dd@18°C Empty coverslip 6 (4) 30000 (13000 cells/cm2) Yes Yes 7dd@20°C 17700 (3) 7 (4) 60000 (26000 cells/cm2) Yes Yes 7dd@20°C 46500 (3) 8 (5) 5000 (2100
cells/cm2) Yes Yes
3dd@37°C + 9dd@37°C Empty coverslip 9 IBIDI Coverslip (no. 10501) 1000 (400
cells/cm2) Yes Yes
3dd@37°C + 6dd@37°C
60000 (3)
10 5000 (2100
cells/cm2) Yes Yes
3dd@37°C + 6dd@37°C 184000 (3) 11 10000 (4200 cells/cm2) Yes Yes 3dd@37°C + 6dd@37°C 212000 (3) 12 20000 (8400 cells/cm2) Yes Yes 3dd@37°C + 6dd@37°C Mould contamination 13 (4) Thermanox Nunc 174934 50000 (22000 cells/cm2) Yes Yes 5dd@20°C 142000 (3)
99 14 (4) Thermanox Nunc 174934 50000 (22000 cells/cm2) Yes Yes 5dd@20°C + 6dd@37°C 142500 (3) 15 (4) 50000 (22000 cells/cm2) Yes Yes 5dd@ 20°C + 6dd@37°C 11000 (3) 16 (4) 50000 (22000 cells/cm2) Yes Yes 5dd@ 20°C + 6dd@37°C Almost empty coverslip 17 (4) 50000 (22000 cells/cm2) Yes Yes 5dd@ 20°C + 6dd@37°C Almost empty coverslip 18 (4) 50000 (22000 cells/cm2) Yes Yes 5dd@ 20°C + 6dd@37°C Almost empty coverslip 19 (4) 50000 (22000 cells/cm2) Yes Yes 5dd@ 20°C + 6dd@37°C Empty coverslip 20 (4) 5000 (2100 cells/cm2) Yes Yes 7dd@ 20°C 250 (3) 21 (4) 5000 (2100 cells/cm2)
Yes Yes 7dd@ 20°C Empty coverslip 22 (4) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C 3750 (3) 23 (4) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C + 6dd@37°C 625 (3) 24 (4) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C 21500 (3) 25 (4) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C + 6dd@37°C 150000 (3) 26 (4) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C 12000 (3) 27 (4) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C + 6dd@37°C 300 (3) 28 (4) 30000 (13000 Yes Yes 7dd@ 20°C + 6dd@37°C Bacterial contamination.
100 (1) 2% gelatin-coated coverslips.
(2) 9-days experiments require 2 medium replacements after the activation, 6-day experiments requires only 1 medium refresh.
(3) Cell number and viability determined by manual counting, with Burker chamber, trypsinized and PBS-washed cells after Trypan Blue staining.
(4) ‘Ice bricks’ heated at 37°C were placed along with the EUs inside a polystyrene box in order to keep them warm.
(5) Technical problems with the EUs occurred (shafts did not work properly and medium replacement did not occur).
Culture support
Both Thermanox and IBIDI coverslips show proper flexibility, resistance and allow a very good cell attachment and proliferation, since they are culture-treated on one side for enhanced cell attachment and growth.
However, Thermanox Coverslips are not suitable for fluorescent microscopy, whereas IBIDI Coverslips show excellent optical characteristics.
Coating
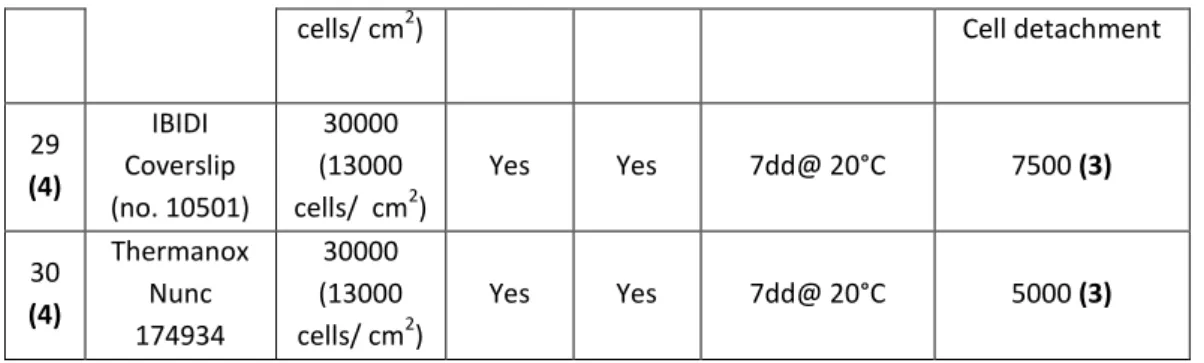
HMEC-1 cells have exhibited a better attachment to coverslip surface when the substrate was coated with 2% gelatin for 20 minutes at RT, independently from the type of coverslip used. After several days from seeding, there were many more cells on coated coverslips than on uncoated ones. In addition, cells on gelatin-coated coverslips look very well spread (Fig. 23.B).
cells/ cm2) Cell detachment
29 (4) IBIDI Coverslip (no. 10501) 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C 7500 (3) 30 (4) Thermanox Nunc 174934 30000 (13000 cells/ cm2) Yes Yes 7dd@ 20°C 5000 (3)
101
This is the reason why only coated coverslips have been used for all the cell growth tests inside the bioreactor (Tab.2).
Initial cell number
The initial number of cells to be seeded on the coverslip surface is one of the most crucial parameter to establish.
In fact, we deduced from the data listed in the table above (Tab.2) that seeding too few cells very probably will lead to their death after the pre-incubation at suboptimal temperature, whereas if there are too many cells on the coverslip an endothelial cell layer will form and easily detach from the culture support surface. To avoid these unwanted events, the initial cell number must be carefully calibrated on the basis of the pre-incubation period of variable duration at suboptimal temperature, the following 6-day incubation at 37°C and HMEC-1 doubling time.
As we can see from experiment # 25 (Tab.2) , by seeding at least 30.000 cells ( a
cell density of 13000 cells/ cm2) most of HMEC-1 will survive during the
incubation at 20°C and can continue to grow at 37°C. On the contrary, a starting number of 50.000 cells (22000 cells/cm2) turned out to be excessive: in fact, during the 5- day incubation at 20°C HMEC-1 have surprisingly continued to proliferate (e.g. experiment #14) and at the end of the 6-day incubation at 37°C no
A B
Figure 23. Leica Microscope, 10x objective. HMEC-1 seeded on uncoated IBIDI
102
more cells were found on the coverslip; most likely ECs formed a thick cell layer that detached as soon as the liquid flow injected inside the culture chamber reached the coverslip.
Incubation protocol
Various incubation protocols have been tested, in terms of temperature and duration of the incubation.
For what concerns the pre-incubation at suboptimal temperature, what we observed was that keeping the bioreactors at temperature below 20°C is not acceptable to preserve live cells, as shown in Tab.2 (experiments #2-5).
On the contrary, cells seeded inside the experimental units (EUs) can be kept at 20°C for up to 7 days, which is the time required for pre-launch activities plus 2 days for orbital flight, within a possible Dragon mission scenario, plus 2 days for a possible delay. After this period, cell number results substantially reduced but it is still acceptable for further growing at 37°C so that, at the end of the 6 day-incubation, the coverslip is at confluence and enough cells can be recovered for further processing.
But HMEC-1 have been incubated at 37°C inside the EUs even for longer periods of time (e.g. 9 days). During a 6-day experiment two medium exchanges are provided, then cells are washed and fixed properly, whereas a 9-day experiment would require three medium exchanges. In this case, there would be a higher risk of cell overgrowth on the coveslip, followed by their death and detachment. Then, we considered worthwhile to supplement MBCD 131 culture medium with 20 mM HEPES buffer, in order to maintain a physiological pH.
103
4.2 Fixing and Storing protocol
In order to select the most suitable fixation and storing protocol for further processing, samples have been subjected to many different fixation tests too. A common reagent for cell fixation is Paraformaldehyde, because it is able to preserve cell structure in an optimal way. But due to safety reasons on board of the ISS, Paraformaldehyde must be replaced by non-harmful chemicals, such as NotoxHisto (EarthSafe Industries, IL USA) and RNA Protect (Qiagen).
For this reason, 4% Paraformaldehyde has been used as a gold standard fixative, to be compared with the performances of the other two commercial reagents.
NotoxHisto
Different solutions of NotoxHisto were tested to fix HMEC-1: the undiluted reagent (as recommended by EarthSafe Industries), NotoxHisto diluted 1:2, 1:5 or 1:10 in 1X PBS.
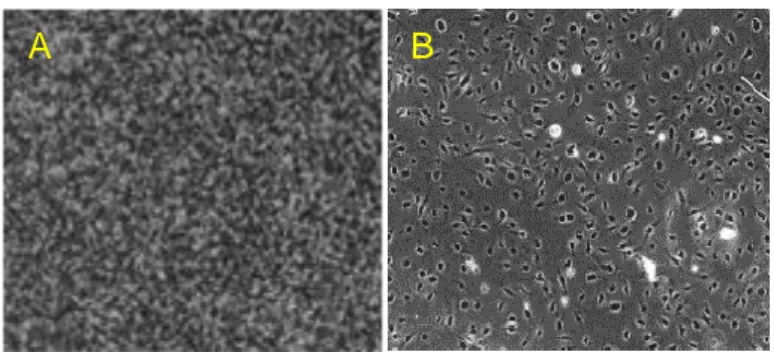
As we can see in Fig. 24, cells fixed with undiluted NotoxHisto look shrunk and are surrounded by a lot of debris (frame B).
Being the fixative too aggressive, we tried to dilute it in 1X PBS buffer and its performance remarkably improved.
In fact, by comparing frame C with A (Fig.24) it is possible to notice that 1:2 NotoxHisto-fixed HMEC-1 look very similar to Paraformaldehyde-fixed cells. On the contrary, fixing with 1:5 NotoxHisto is suboptimal, because cells look folded and partially detached, especially after washing them with PBS (not shown). Additional dilution of the fixative (1:10 NotoxHisto) is far too high, cells are folded and scarce, many detached after washing.
For what concerns the storing protocol, HMEC-1 have been stored at 4°C in both 1X PBS and NotoxHisto; in the latter situation, though, there is the risk that cells get over fixed.
104
RNA Protect
RNA Protect has been selected to stabilize nucleic acid structure; initially we tested the undiluted reagent on a confluent coverslip and observed cell detachment as soon as the fixative contacted HMEC-1. An analogous result can be obtained by diluting the reagent 6:1 in culture medium, as suggested by Qiagen.
Therefore, in order to avoid cell detachment, the dilution ratio needs to be overturned: we used RNA Protect diluted 1:6 in culture medium (Fig. 25.A) and in 1X PBS (Fig. 25. B). In the former solution many aggregates seem to sediment on cells (frame A); the latter one, on the contrary, allows to obtain a satisfying result. A C B D E
Figure 24. Leica Microscope, 10x objective. HMEC-1 fixed in
: A) 4% PFA B) NotoxHisto undiluted C) 1:2 NotoxHisto D) 1:5 NotoxHisto E) 1:10 NotoxHisto.
105
Also in this case two storing solutions, 1X PBS and RNA Protect, have been tested. Anyway, in order to preserve nucleic acid integrity in the best possible way, storing HMEC-1cells in the same fixative solution is much more recommended.
4.3 Genomic DNA extraction
The DNeasy Blood & Tissue Kit (Qiagen) allowed a good purification of the total genomic DNA.
To quantify the extracted material we performed NanoView™ Spectrophotometer (GE Healthcare) measurements and we obtained the following concentrations:
First elution : 7 ng/µl x 100 µl = 700 ng Second elution : 6,6 ng/µl x 100 µl = 660 ng
The total yield can be easily calculated and amount to 1,36 µg of gDNA.
A B
Figure 25. Leica Microscope, 10x objective. Cells fixed with
RNA Protect diluted in: A) Medium, 1:6 and B) in 1X PBS, 1:6.
106
By using protocols adapted for low-amount materials (Taiwo et al., 2012), it is possible to perform a methylome analysis starting from such low DNA concentrations.
To assess the integrity of the DNA, samples were loaded as follows to a 0,6% agarose gel in 1X TBE:
1) 1:10 (DNA diluted in TE) 2) not diluted DNA
10 µl + 2 µl of 6x Orange G 10 µl + 2 µl of 6x Orange G
first elution
3) 1:10 (DNA diluted in TE) 4) not diluted DNA
10 µl + 2 µl of 6x Orange G 10 µl + 2 µl of 6x Orange G
second elution
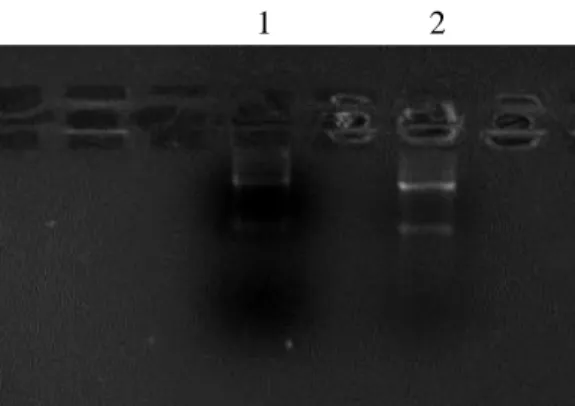
As we can see in the figure below (Fig.26), the purified genomic DNA is intact and its integrity has been preserved during the extraction procedures.
I elu II elu
1 2 3 4
Figure 26. Agarose gel image. Lane 1 and 2 are
loaded with first elution sample, lane 3 and 4 with second elution.
107
4.4 Total RNA extraction
The RNeasy Mini Kit (Qiagen) allowed the purification of the total RNA starting from one 90% confluent coverslip, which amounts to 90.000 cells approximately. The concentration of the extracted material was measured at the NanoView ™ Spectrophotometer (GE Healthcare):
First elution (30 µl) 13,6 ng/µl Second elution -2,6 ng/µl
To obtain the total yield we have to multiply the RNA concentration only by the volume of the first elution:
Total yield: 13,6 ng/µl x 30 µl = 408 ng
Concluding, 408 ng of total RNA were purified from one single confluent coverslip and are enough for a trascriptome analysis, since nowadays high-sensitivity methods that use only 100 ng of RNA input are available on the market.
To assess the integrity of the RNA, samples were loaded as follows to a 2% agarose gel in 1X TBE:
7 µl of tRNA (4 µl of RNA from I elution + 3 µl of RNA from II elution) + 7 µl of 2X Loading Buffer
1) about 10 µl of RNA sample + Loading Dye 2) about 4 µl of RNA sample + Loading Dye
108
4.5 Morphological analysis
After fixing and storing, it is of fundamental importance that cells maintain their morphological characteristics unaltered.
In order to evaluate this, actin cytoskeleton and myosin have been stained after different periods of storing. Once again, Paraformaldehyde-fixed cells were considered a reference point in the analysis of cell morphology.
4.5.1 Short-term storage
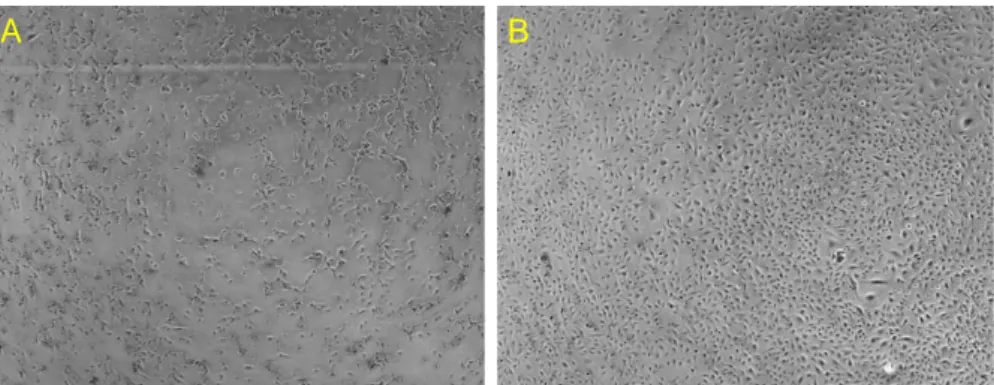
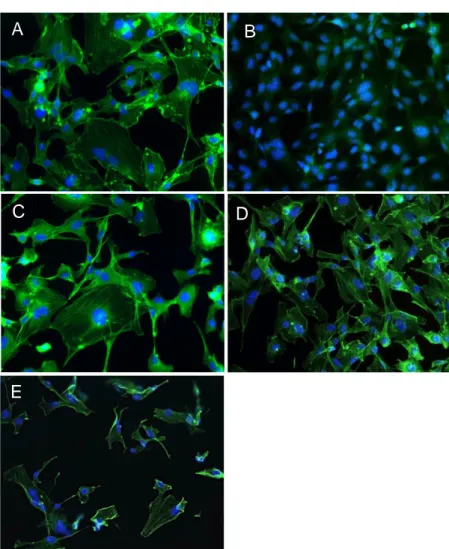
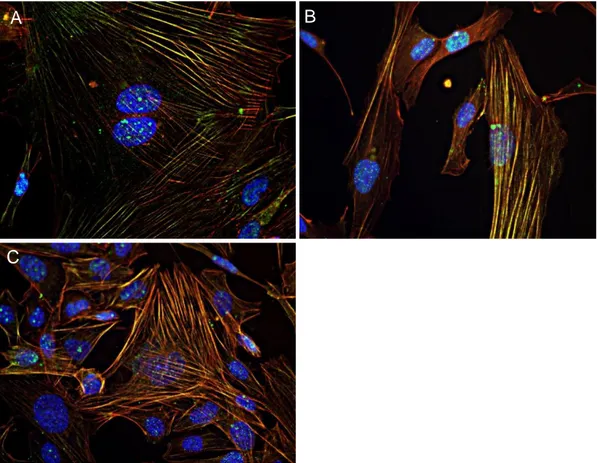
The photo panel in Figure 28 concerns actin staining with Phalloidin 488 (Cytoskeleton) after 4 days from fixation. From top to bottom, clockwise: HMEC-1 were fixed with 4% Paraformaldehyde (A), undiluted NotoxHisto (B), HMEC-1:2 NotoxHisto (C), 1:5 NotoxHisto (D) and 1:10 NotoxHisto (E).
Figure 27. Agarose gel image. The two lanes
are loaded with the same sample. Lane 1 contains a smaller amount of RNA sample.
109
It is obvious that undiluted NotoxHisto is not able to preserve cell architecture, since actin fibres are not labeled at all, whereas when diluted 1:2 in PBS actin staining remarkably improves. In fact, the result is comparable to the one obtained with Paraformaldehyde.
On the contrary, cells look folded and partially detached when fixed with more diluted solutions of NotoxHisto (frames D and E), since the fixing methods are
B
E
D A A
C
Figure 28. Nikon Ti, 20x objective. Actin staining (Phalloidin 488,
Cytoskeleton) after 4-day storage: HMEC-1 fixed with A) 4% PFA, B) NotoxHisto undiluted, C) 1:2 NotoxHisto, D) 1:5 NotoxHisto and E) 1:10 NotoxHisto. Nuclei in blue (DAPI).
110
not optimal and cause the detachment of many cells during the staining procedures.
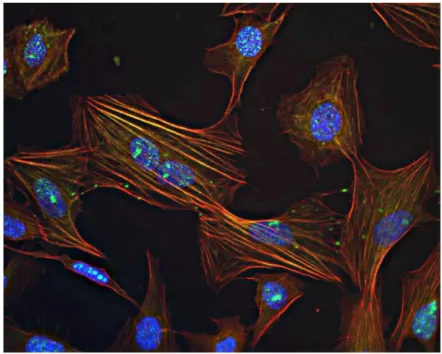
As we can see in the figure above (Fig.29, frame B), staining of 1:2 NotoxHisto-fixed cells is still successful, even after 1-month storage in PBS at 4°C: actin fibres are well defined and cell architecture remains unaltered.
In order to evaluate whether protein antigens were preserved as well, in addition
to Phalloidin staining (in red, SIGMA), we performed indirect
immunocytochemistry against phospho-myosin (Anti-PMLC2, Cell Signaling Technology), as shown in Fig.30.
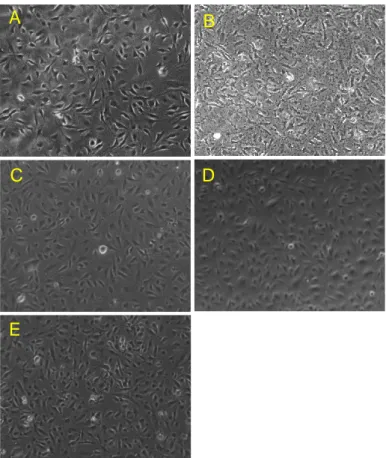
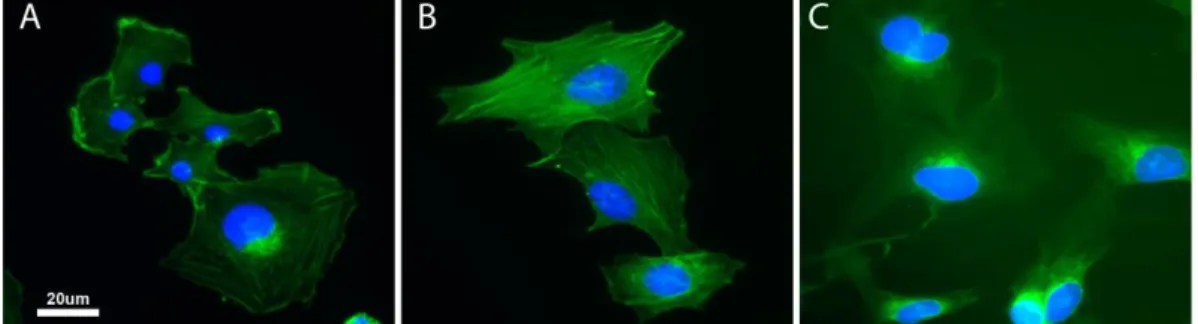
In this case, fixed HMEC-1 cells were subjected to staining procedures at different times: in frame A and B, respectively, Paraformaldehyde and 1:2 NotoxHisto-fixed cells were stained soon after their fixation, without being stored
at 4°C and served as controls; whereas HMEC-1 in frame C have been stored at
4°C for more than 40 days, before being subjected to immunocytochemistry. By analyzing the obtained results, 43-day stored cells show an excellent staining if compared to the fresh samples; the cytoskeleton is kept perfectly intact, since actin fibres appear to be well delineated and the myosin labelling is associated with them to perfection.
Figure 29. Nikon Ti, 40x objective. Actin staining (Phalloidin 488, Cytoskeleton) after 1-month
storage: HMEC-1 fixed with A) 4% PFA, B) 1:2 NotoxHisto and C) NotoxHisto undiluted. Nuclei in blue (DAPI).
111
4.5.2 Long-term storage
At the end of the experiment in Space, several months could be required from sample download to handover to science team. Therefore, it is crucial to perform cytostaining and immunocytochemistry on long-term stored samples.
In order to evaluate whether protein antigens were preserved and cell morphology retained after a long-term storage of 150 days, we performed actin staining and
A B
C
Figure 30. Nikon Ti, 40x objective. Immunostaining showing actin fibres in red (Phalloidin-Atto
550, SIGMA), phospho-myosin light chain 2 in green (Anti-PMLC2, Cell Signaling Technology) and nuclei in blue (DAPI).
A) 4% PFA-fixed cells, B) 1:2 NotoxHisto-fixed cells, C) 1:2 NotoxHisto-fixed cells stored in PBS at 4°C for 43 days.
112
indirect immunocytochemistry against phospho-myosin (Anti-PMLC2, Cell Signaling Technology), as shown in Fig.31.
The figure refers to 1:2 NotoxHisto-fixed HMEC-1 cells that have been stored at 4°C for 5 months in 1X PBS, before being subjected to staining procedures.
We can see that the co-labeling of Phalloidin and phospho-myosin persists and that cells do not undergo morphological alterations even after such an extended period of time from fixation.
Lastly, fluorescent staining of RNA Protect-fixed cells has been tested as well but was not successful at all (not shown), because the fixing method is not suitable for preserving cell architecture and it will not be used for this purpose.
C
Figure 31. Nikon Ti, 40x objective. Immunostaining showing actin
fibres in red (Phalloidin-Atto 550, SIGMA), phospho-myosin light chain 2 in green (Anti-PMLC2, Cell Signaling Technology) and nuclei in blue (DAPI). 1:2 NotoxHisto-fixed cells that have been stored in 1X PBS at 4°C for 150 days.
113
5. CONCLUSIONS
At this preparatory phase of the SFEF project, different conditions for the implementation of the experiment in Space have been tested, focusing on 1) designing the protocol for cell culturing, fixing and storage inside the bioreactor; 2) testing several pre-flight incubation protocols to simulate different mission scenarios; 3) evaluating the suitability of fixed samples for subsequent experimental procedures.
In order to develop optimal culturing conditions inside the bioreactor, different cell culture supports, starting cell numbers and incubation protocols have been evaluated.
Then, highly confluent coverslips were used for nucleic acid extraction to make sure that they could yield enough total RNA and genomic DNA to perform respectively transcriptome and methylome analysis, with protocols adapted for low-amount materials.
Due to safety reasons and hardware design, instead of the gold standard fixative 4% paraformaldehyde, several other fixative solutions have been tested for cell immunofluorescence to be performed with various markers.
For a general quality control of cell morphology and cytoskeleton architecture after storing for different periods of time, fixed cells were stained also with Phalloidin.
The results obtained have been used to write a scientific report for ESA, the ESR document, which contains the rationale and all the experimental details for the SFEF mission.
This chapter is divided into several sections concerning cell growth inside the bioreactor, fixing and storing protocols of the samples and their morphological analysis; each section will describe in detail all the parameters established for the implementation of the experiments to be performed inside and outside the bioreactors.
114
5.1 Cell growth inside the bioreactor
Culture support
Both Thermanox and IBIDI coverslips allow a very good cell attachment and proliferation, beyond offering a proper flexibility. However, Thermanox Coverslips had a great limitation for our studies that is their unsuitability for fluorescent microscopy; fortunately, the COC polymer of IBIDI Coverslips has permitted to overcome this problem, thanks to its excellent optical characteristics. Thus, cells seeded on Thermanox Coverslips will be used to extract nucleic acids, whereas fluorescent immunocytochemistry will be performed on IBIDI Coverslips-grown cells.
Coating
HMEC-1 cells are seeded on 2% gelatin-coated coverslips, since they have exhibited a better attachment to coverslip surface.
It is noteworthy to remember the importance of cell adhesion for the success of the experiment inside the bioreactor; in fact, even though flow rates of medium and fixative have been carefully evaluated in order to avoid the onset of shear stress on the cells, they still have to adhere to the culture support in the best possible way.
Starting cell number
Another crucial parameter is represented by the initial number of cells to be seeded on the coverslip surface. In effect, different factors have to be taken into account in order to establish the starting cell number: the pre-incubation period of variable duration at suboptimal temperature, the following 6-day incubation at 37°C and HMEC-1 doubling time.
115
In the end, what came from the tests on cell seeding density is that an intermediate confluency of 30% ( approximately 30.000 cells) is required for an adequate cell growth inside the bioreactor.
In fact, a density of 13.000 cell/cm2 allows a relatively high percentage,
approximately 40% on average, of HMEC-1 to survive during the incubation at 20°C, so that their number is still acceptable for further growing at 37°C.
Pre-incubation
Based on the results obtained so far, we conclude that cells seeded inside the experimental units (EUs) can be kept at 20°C for up to 7 days, which is the time required for pre-launch activities plus 2 days for orbital flight, within a possible Dragon mission scenario, plus 2 days for a possible delay.
After this incubation period at suboptimal temperature, cell number results substantially reduced but it is still acceptable for further growing at 37°C so that, at the end of the 6 day-incubation, the coverslip is at confluence and enough cells can be recovered for further processing.
On the contrary, keeping the bioreactors at temperature below 20°C is not acceptable to preserve live cells.
Incubation at 37°C
HMEC-1 cells are able to grow and proliferate inside the culture chamber of the bioreactor, when incubated at 37°C for 6 days.
Being the culture chamber a completely isolated environment, MBCD 131 culture medium is supplemented with 20 mM HEPES buffer, in order to maintain a physiological pH despite changes in carbon dioxide concentration.
During the 6-day experiment two medium exchanges are provided (once every three days), then cells are washed and fixed properly.
116
5. 2 Fixing and storing protocol
Fixative
Depending on the application the sample is committed to, fixation protocol varies: if nucleic acid purification is meant to be performed, cells are washed for 1 minute with 1X PBS in order to remove all traces of culture medium that may interfere
with the fixative solution andfixed for 1 hour at 37°C in 1:6 1X PBS-diluted RNA
Protect (Qiagen).
On the contrary, those samples destined to fluorescence staining are washed with 1:2 1X PBS-diluted NotoxHisto (EarthSafe Industries, IL USA) and fixed in the same solution for 1 hour at 37°C.
These are the best working dilutions for both the fixatives; sample stabilization, fixation quality and cell morphology preservation are excellent under these conditions.
The two different protocols are schematized below, indicating all the fluid injections (Fig. 31 and 32) that are meant to be performed during a 6-day experiment inside the EUs.
Storing
Once the material has been fixed, cells need to be stored inside the experimental units until their unloading, which might take months.
To preserve the nucleic acid integrity, RNA protect-fixed cells are stored in a fresh aliquot of the same fixative solution (1:6 RNA Protect) at 4°C.
On the other hand, samples fixed in NotoxHisto are stored in 1X PBS at 4°C to avoid over-fixation of the sample, which may cause an altered cell morphology.
117
5.3 Nucleic Acid extraction
By using purification kits that are suitable for smaller samples, we extracted the
genomic DNA and the total RNA starting from single coverslips (2 x 1.05 cm2,
Thermanox Nunc 174934) at 90% confluency (about 90.000 cells).
RES#1 Medium RES#2 Medium RES#3 PBS RES#4 RNAProtect RES#5 RNAProtect 1st Medium IN 2nd Medium IN PBS IN RNA Protect IN RNA Protect IN Culture Chamber with Medium
Beginning of the experiment
Culture Chamber with Fixative
End of the experiment
RES#1 Exhausted RES#2 Medium ReS#3 RES#4 Exhausted PBS RES#5 Exhausted RP
Initial Medium OUT 1nd
Medium OUT
2nd Medium OUT PBS OUT
RNA Protect OUT
Figure 32. RNA Protect-fixed samples fluidic scheme. RES= Reservoir.
RES#1 Medium RES#2 Medium RES#3 NotoxHisto RES#4 NotoxHisto RES#5 PBS 1st Medium IN 2nd Medium IN PBS IN NotoxHisto IN Culture Chamber with Medium
Beginning of the experiment
Culture Chamber with Fixative
End of the experiment ReS#4 Exhausted NH
RES#5 Exhausted NH Initial Medium OUT 2nd Medium OUT NotoxHisto OUT NotoxHisto OUT
Figure 33. NotoxHisto-fixed samples fluidic scheme. RES= Reservoir.
NotoxHisto IN 1st Medium OUT RES#1 Exhausted RES#2 Medium ReS#3
118
In both cases, the total yield of the purified material is estimated to be enough for future experiments, such as the analysis of the transcriptome and of DNA methylation, following protocols adapted for low-amount materials.
5.4 Morphological Analysis
In order to evaluate whether cell morphology is retained after variable periods of time from fixation with 1:2 NotoxHisto, HMEC-1 cells have been subjected to staining of the actin cytoskeleton.
In addition to it, also myosin have been marked with a specific antibody; in this way, beyond the general cytoskeleton architecture, it has been possible to evaluate whether even antigens were preserved as well.
Based on the results obtained so far, we proved that fluorescent staining of actin fibres is still successful, even after 1 month from fixation; in the meanwhile coverslips have been stored in 1X PBS at 4°C. Cell morphology and cytoskeleton architecture are well preserved, nuclei are round and regular and actin fibres are very well defined, compared with morphologic controls.
Furthermore, from the results of fluorescent immunocytochemistry against phospho-myosin, we can conclude that both protein antigens and post-transcriptional modifications are kept intact even during a storing time of 150 days. This result is significantly important, given that several months after sample download could be necessary until handover to science team.
119
The table below summarizes all the experimental parameters established during the preparatory work of the SFEF project and described in the previous sections (Tab.3).
All these parameters have also been used to compile an official document for the European Space Agency, the Experiment Scientific Requirements (ESR).
Application Coated coverslip Initial cell number Incubation protocol Washing buffer Fixative solution Storage Nucleic acid extraction Thermanox Nunc 174934 30.000 (13.000 cell/cm2) 7dd@20°C + 6dd@37°C 1X PBS RNA Protect 1:6 RNA Protect 1:6 Fluorescent staining IBIDI Coverslip (no. 10501) Notox 1:2 Notox 1:2 1X PBS
Table 3. Summary table, indicating the different parameters selected depending on the
120
6. Future Perspectives
The SFEF project aims at a better understanding of the modulation of endothelial functions in Space, since endothelial cells are responsible for the integrity of the vascular wall.
Although it has not been formally assigned yet, a possible launch date is foreseen for the end of 2015.
We hope SFEF mission will permit to determine and distinguish the biological effects on microvascular endothelium of both microgravity and cosmic radiations. Therefore, a short-term expected outcome is to identify possible markers of Space flight-induced endothelial suffering, such as cell senescence and DNA damage.
It is worth to carry out this study, since its results may contribute in the long run to define new prevention, rehabilitation and therapeutic protocols for astronauts returning to Earth. Especially within sight of long-term Space missions, it will be necessary to provide potential countermeasures to prevent cardiovascular deconditioning in astronauts.
But Space Biology does not limit itself to affect Space crews’ lives, in fact common persons suffering from inflammatory, degenerative and cardiovascular pathologies might benefit too from its possible outcomes.
Microgravity represents indeed a unique model to simulate, in a very accelerate way, all those events that normally occur during the aging process on Earth or that are a consequence of sedentary life.
So, this kind of study might offer new insights also in elucidating the mechanisms of tissue and organs impairment, due for example to the passing of time, and in finding potential treatments and preventive strategies.
In addition, it is also possible to exploit a microgravity environment to produce tissue equivalents for a variety of basic and applied medical purposes (Unsworth
121
et al., 1998). Analogously, by generating tumor spheroids, there might be repercussions on the understanding of cancer development (Marrero et al., 2009).
In conclusion, investigating how Space flight affects endothelial function is a promising basic biological research that might influence not only Space mission duration and crews’ health, but also general public’s lives.