Mary
AnnLiebert,
Inc., PublishersBiologic
and Molecular
Characterization
of Producer
and
Nonproducer
Clones from HUT-78 Cells
Infected with
a
Patient
HIV
Isolate
MAURIZIO
FEDERICO,1
FAUSTO
TITTI,1
STEFANO
BUTTÓ,1
ANGELA
ORECCHIA,1
FRANCESCA
CARLINI,1
BRUNELLA
TADDEO,1
BEATRICE
MACCHI,2
NICOLA
MAGGIANO,3
PAOLA
VERANI,1
and
GIOVANNI B.
ROSSI1
ABSTRACT
HUT-78 cells
wereinfected with
a reversetranscriptase (RT)-positive
supernatant
of
acul-ture
of
peripheral
blood
lymphocytes
(PBL)
from
anAIDS
patient
and then cloned. Of
these
clones,
twohave
been isolated and
characterized.
Clone DIO is
persistently
and
pro-ductively
infected
with
anHIV variant. The clone
F12,
in
spite
of
the presence of
aninte-grated
full-length
HIV
provirus,
does
notrelease virus
particles
in the medium.
DIO and F12 clones
substantially
differ in
termsof
protein
pattern;
that
is,
DIO is
super-imposable
toinfected HUT-78
cells,
whereas F12 exhibits
adecreased
uncleaved
p55
gagprecursor and the presence of uncleaved
«pi60
and of
aunique pl9,
although they
do
notshow
qualitative
orquantitative
differences in viral RNA
synthesis.
Restriction
patterns
of
F12
proviral
DNA
do
notshow
major genomic
deletions. These
results
indicate that
F12
clone cells carry
anHIV genome with minor mutations that
probably
affect the
correctproduction
of viral
proteins
at aposttranscriptional
level.
Inaddition,
the F12 clone is
resistant
tohigh-multiplicity superinfection
with HIV-1
orHIV-2.
INTRODUCTION
A
peculiar feature of human immunodeficiency virus(HIV),
theetiologic
agent
of theacquired
immunodeficiency syndrome
(AIDS)
and associated clinicaldisorders,
is thebiologic
andgenetic
heterogeneity
among different isolates.'Laboratory
ofVirology,
IstitutoSuperiore
diSanità, Rome,
Italy.
department
ofExperimental
Medicine, 2ndUniversity
ofRome, TorVergata,
Rome,Italy.
'Department
ofPathology,
Université Cattolica del S. Cuore, Rome,Italy.
Since 1985 wehave
performed
alarge
number of HIV isolations frompatients
withAIDS orAIDS-re-lated
complex.1
Preliminary
characterizationsuggested
that isolates varyboth interms ofbiologic
behavior(i.e.,
differentialability
toproductively
infect humancells)
and of structuralcomponents
(i.e.,
viral pro-teins and restrictionpatterns
ofproviral
DNA).
These results are inagreement
with data from differentlaboratories. Evanset
al.2
foundthatvariousCD4+
human cellsaredifferently
sensitivetothe infectionby
thesame HIVisolate;
at thesametime,
different isolates may ormay notinfect the same celltype.
Alsogenomic variability
of HIVisolates,
analyzed
via DNA restrictionpatterns3-7
orby comparing
these-quences of differentcloned
HIVs8
has beenreported.
Taken
together,
these data demonstrate that HIV isolates varyremarkably
in nucleotide sequence andconsequently
inprotein composition.
Themostvariable sites are located in theenvgene, which codes forthe
envelope proteins,
andmoreprecisely
inaregion coding
fortheextracellularportion
of HIVgpl20.9-10
Biologically,
thegenomic variability
ofHIV,
alsotypical
of otherlentiviruses,
such as theequine
infec-tious anemia virus
(EIAV),
canplay
a role infighting
the host immune defense mechanismsduring
the courseofinfection.1112
The
great
variability
ofHIVposes thequestion
of whetherit ispossible
todetect in the samesubject
thesimultaneous presence of two or more
genotypically
distinctHIVvariants,
derived eitherby
an in vivosuperinfection
with different HIVs orby
the de novoformation ofHIV variantsduring
thecourse ofHIVinfection. This
possibility
was raisedwhen the presence oftwocomplete
genomic equivalents
wasrecog-nized
by HIV-specific
probes
both intheDNA of H9/HTLV-IIIB cells(originally
infectedwith amixtureof HIV
isolates)13
and in the DNA of cells infected withasingle
HIVisolate.1>3'5
Morerecently, Koyanagi
et
al.14
have isolated two related butgenotypically
distinct variants of HIV from two different sources(brain
tissueandcerebrospinal
fluid)
of thesamepatient.
Inaddition,
Von Bliesenetal.15
have isolated andmolecularly
cloneddifferentgenetic
variants of HIV fromperipheral
bloodlymphocytes
(PBL)
ofasingle
patient.
Inour
laboratory
we have infected HUT-78 cultures witha reversetranscriptase (RT)-positive
superna-tantof PBL fromanAIDSpatient.
Asubsequent
cellularcloning by
thelimiting
dilution methodgave riseto several
clones,
two of which are characterized here. One clone releases HIVvirions,
and the otherexhibits HIV markers but does notrelease
RT-containing
virions orviralproteins
and cannotbesuperin-fected
by
other HIVisolates.MATERIALS
AND
METHODS
Cell cultures and
cloning
Separation,
stimulation,
and cultivation ofPBLfrom AIDS donorswere describedelsewhere.1
Allcellcultures
(uninfected
HUT-78,
DIO,
and F12clones,
HTLV-IIIB-infectedHUT-78)
were grown in RPMI-1640 medium(Flow
Laboratories, Scotland)
containing
20% inactivated fetal calfserum(FCS)
andsplit biweekly.
Cellcloning
wasperformed
by
thelimiting
dilution method in96-wellplates.
HIV
infection
and detection
HUT-78
cells,
previously
treatedwithpolybrene
(1
p-g/ml; Sigma,
St.Louis, MO),
were infectedby
anultracentrifuged
(200,000
x gfor 30minutes)
RT-positive
supernatant
(about 10,000
cpm per106
cells).
Virus was absorbed for 1 hour at37°C
with occasionalshaking,
and then cellswereresuspended
incom-plete
medium andsplit
at the concentration of 3 x105
per ml twicea week. The presenceofHIV in thesupernatant
was monitoredby
reversetranscriptase
assay asdescribed,1
and/orby antigen
capture
assay(Cellular
ProductsInc., Buffalo, NY).
Electron
microscopy analysis
Cellswere
prefixed
insuspension
with 2%glutaraldehyde
in 0.1 Mphosphate
buffer,
pH
7.4,
for 1 hour1% osmium tetroxide for 45
minutes,
dehydrated
ingraded
ethanol,
and embedded inEpon-812.
Thin sections werestained withuranyl
acetateand leadcitrateand examined inaPhilips
EM400transmissionelectron
microscope.
Indirect
immunofluorescence
assay(IFA)
The presence of
cytoplasmic
HIV-relatedantigens
was assessedby
IFA asdescribed.16
Briefly,
cells werewashed twicewith andresuspended
in PBS at106
per ml. Cellsuspension
(20
pi)
wasspotted
ontoaslide,
airdried,
andfixedincoldacetonefor 10minutes atroomtemperature.
Patient'sserumdiluted 1:20(20
p,l)
wasapplied
onthe fixed cells and incubated for 45 minutes atroomtemperature.
The slides werethen washed for 1 hour in
PBS,
and 20p,l
of 1:20 rabbit antihumanIgG
conjugated
with fluoresceinisothiocyanate
(FITC,
Cappel
Lab., Cochranville,
PA)
was added and incubated at 37°C for 60minutes.The slides werewashed
extensively
in PBS beforemicroscope
examination underultraviolet(UV)
illumi-nation. Uninfected HUT-78 cells were usedasnegative
control.To assess the presence of CD4 membrane
antigen
and ofHIV-specific
membraneantigens,
weper-formed the membrane indirect immunofluorescence as
described.17
Briefly,
5 x105
cells were washedthree timesin PBS
plus
5%FCS,
thepellet
wasdried,
and 10pj
ofappropriately
dilutedanti-CD4mono-clonal orof 1:20 diluted
patient's
serum wasadded andincubated for 20minutes. Cells werethen washed three times in PBSplus
5%FCS,
and 10pJ
of either 1:20 rabbit antimouseF(ab')2
or 1:20 rabbitanti-human
IgG, conjugated
withFITC,
wasaddedatroomtemperature.
Appropriate
cellnumbers werespotted
ontoa
slide,
airdried,
andfixedin ethanol-acetic acid(9:1)
for20 minutesat—
20°C,
andthenrehydrated
and washedthree
times in PBS before UVmicroscope
examination.Radioimmunoprecipitation
assay(RIPA)
HIV-infected and uninfected HUT-78 cells were seeded at 1.5 x
106
per ml and labeled with 100p.Ci/ml
of[35S]cysteine
(NEN-DuPont, Boston, MA,
specific activity
1008Ci/mmol)
for 6 hours after 2 hours of starvationincysteine-free
medium. For32P
labeling,
HTLV-IIIB-infected HUT-78 and F12cellswere seeded at
106
perml inphosphate-free
medium and afteranovernight
incubation labeled with 200p-Ci/ml
of[32P]orthophosphoric
acid(NEN-DuPont,
specific activity
3000Ci/mmol)
for 6 hours. Thecellpellet
wasresuspended
inradioimmunoprecipitation
(RIPA)
buffer(50
mM TrispH
7.4,
0.5% TritonX-100,
100 mMNaCl,
and 100U/ml ofaprotinin).
Thepostnuclear
supernatant
containing
about4 X106
cpm was
exposed overnight
toprotein-A Sepharose
(Pharmacia,
FineChemicals, AB,
Uppsala,
Sweden)
previously
incubated with apositive
reference human serum. After three washes in RIPA buffer and onewash in 10 mM
Tris-HCl,
pH
7.4,
and 0.1 MNaCl,
thepellet
wasresuspended
in 50 u,ldouble-strength
SDS-PAGEsample
buffer(10
mMTris-HCl,
pH
8.0,
2%SDS,
and 2%ß-mercaptoethanol),
boiled 5minutes,
andrun on adiscontinuous 12% or on a 12-20% lineargradient
SDS-polyacrilamide gel,
bothwith 3.5%
stacking gel.
Thegel
wasfixed, stained,
andautoradiographed.
Theexperimental procedure
forpulse-chase experiments
was identical as fortheRIPA,
except
that after 6 hoursoflabeling
the cellswerewashed three times withPBS and
resuspended
at 1.5 x106
perml incoldcomplete
medium. Cellswerethen
collected
andprocessed
as in RIPAat0, 3,
6,
and 9hours after the removal of[35S]cysteine.
Western
blot
(WB)
analysis
Postnuclearcell
lysates
(200
u,g)
werefractionatedby electrophoresis
on apolyacrylamide-SDS
slabgel.
Proteinswere transferredto anitrocellulose sheet
(0.22
p,m;Bio-Rad, Richmond,
VA)
viaelectrophoretic
blotting,
washed,
and blocked with nonfatdry
milkto minimizenonspecific binding.
Nitrocellulosestrips
werereactedwith 1:100 dilution ofa
positive
humanserum. After thewashing cycles,
bound viralproteins
werevisualized
through
aperoxidase-labeled
goat
antihumanIgG (Bio-Rad).
Nucleic acid
analysis
DNA.
High-molecular-weight
DNA was extractedfrom cellclones,
and 15 p,g wasdigested
with theappropriate
restriction enzymesaccording
to the manufacturer's recommendations. The DNA was thensubjected
toelectrophoresis
in 0.8% agarose slabgel
and blottedthrough
ahigh
electric field(Transblot,
Bio-Rad)
ontoGene Screen-Plus filters(NEN-DuPont).
Hybridization
wasperformed
at 42°C in ashaking
water bath for 24 hours in 50%formamide,
1 MNaCl,
5%dextran-sulfate,
1%SDS,
and 100p.g/ml
of sonicated salmon sperm DNA. Filters were thenwashedtwice for 30minutes in
double-strength
standardsalinecitrate(SSC)
atroomtemperature,
twiceindouble-strength
SSC and0.1% SDS at42°C,
andfinally
twicein SSC diluted0.1 x atroomtemperature.
The
probes
were nick translatedat aspecific
activity
of about2 x108
cpm/p,g
DNAusing
a32P-labeled
deoxicytidine triphosphate
(NEN-DuPont;
specific activity
3000Ci/mmol).
Probes utilized werethe
fullgenomic length
(less
180bp
at5'-LTR)
BH10probe (gift
of Dr. R.C.Gallo, Bethesda, MD),
a 3.8 kbgag-pol,
and a 1.9 kb envprobe (gift
ofHoffmann-LaRoche, Basel,
Switzerland)
corresponding
to themost conserved sequences
overlapping
the HIV exones. Filters werefinally exposed
to Kodak SO-282films for 24-72 hours inan holder with
intensifying
screens.RNA. Total RNA was extracted
by
theguanidine-isothiocyanate method18;
thepoly-A+
RNA wasseparated
by oligo-dT
cellulose(Pharmacia)
chromatography.
Poly-A+
RNA(3-5
p,g)
wassubjected
toformaldehyde-formamide denaturing gel electrophoresis
and then blotted andhybridized
asdescribed fortheDNA
procedures.
RESULTS
Isolation and
biologic
characterization
of
D10 and F12 clones
HUT-78 cellswereinfectedwithan
RT-positive
supernatant
of PBL fromanAIDSpatient.
HIV-infectedcultures weremaintained
by adding weekly
fresh HUT-78cells until cellviability
dramatically
decreased.Fresh HUT-78 cells were then infected with a 100-foldconcentrate
supernatant
of the former culture and after 16 passages were clonedby
thelimiting
dilution method. Several HIV-infected cell clones wereobtained,
two ofwhich wereexpanded
and characterized. The first clone(D10)
ischronically
HIVin-fected,
releases HIVvirionsathigh
levels(Table 1),
andreplicates indefinitely
withoutcocultivationwith uninfected cells. The second(F12),
albeitpersistently
HIV infected(Table 1),
doesnotrelease infectious viralparticles
in thesupernatant,
asconfirmedby
the unsuccessful infection ofHIV-susceptible
cells(PBL,
H9,
orHUT-78)
witha500-fold concentratedF12supernatant.
Thepresence ofnoninfectious aberrant HIVparticles
in F12 clonewasruledoutby
electronmicroscopic analysis.
Nomatureorbudding particles
werenoticed in
F12,
whereasmaturetype
Cparticles
wereclearly distinguishable
in D10 cells(data
notshown).
Table 1. BiologicCharacterizationofD10andF12 HIV-Infected CellClones
Cell membraneIFAb
(%)
Cytoplasmic IFA,b
-RT1 HIV
antigen
Syncytia
anti-HIV(%)
anti-CD4 anti-HP/ HUT-78 - — 98.0 — HUT-78/HTLV-HIB + + + 93.6 — NDd D10 + + + 91.0 — 73.4 F12 -92.3 — 1.6 "Mean valuesatconfluence:HUT-78/HTLV-IIIB, 3 x 105cpm/ml;
D10,5 x 105cpm/ml;
HUT-78 andF12,
below thebackground.
bValuesofa
representative experiment.
cAbnormalamountsof cellulardebris in F12
supernatantsmaycause afaint
positivity, just
above thebackground.
Toassessthe
possible
release of viralproteins,
F12cellswerelabeledwith[35S]cysteine,
thesupernatant
was concentrated 500-fold via ultrafiltration(Centricon,
Amicon, Danvers, MA; cutoff, 10,000 daltons)
and
analyzed by
RIPA inparallel
with a similarpreparation
from a D10 clone culture. Evenloading
a10-fold excess of cpm, no viral
products
wererecognized
in F12supernatant,
whereas thetypical
HIVantigens
wereremarkably
present
in D10supernatant
(data
notshown).
The
growth
rateof thetwoclones issimilar,
asboth reachsaturationatabout2 x106
cells permlafter4days
ofculture. Viral infectionandproduction
wereassessedby
RTassay,by
theantigen
capture
assay, andby cytoplasmic
IFAperformed
withapolyclonal
anti-HIVserum. Table 1 shows that thepercentage
ofIFA-positive
cells isequally high
(80-90%)
inF12, D10,
and controlpositive
cells(HUT-78/HTLV-IIIB).
Thetwoclones were alsoanalyzed
for thepresence ofHIV-specific antigens
on theplasma
membrane.A
partially purified
human anti-HIV serumrecognized
73.4% D10 cells andonly
1.6% F12 cells. Thesedata as well as those from RIPA
experiments
(see later)
indicate the presence ofHIV-specific gpl20
antigens
ontheplasma
membrane ofthe D10 cellclonebut notonthat of the F12clone.Finally,
the presence of CD4receptor
sites was testedusing
a monoclonal anti-CD4+antibody.
Asalready
described for otherHIV-infected celllines,19
neither D10 norF12 clones exhibitany CD4mem-brane
antigen.
Allthese features are
invariably
conservedeven after 12 monthsofculture.Attempts
to rescueHIVfrom
F12 clone cellsF12clone cells havebeen treated withoutsuccesswithawide
spectrum
ofdosesoffluoro-, bromo-,
andiododeoxyuridine
forpossible
rescue of theintegrated provirus
(data
notshown).
Further,
wetriedtocomplement possible
defects of F12proviral
genomeby superinfecting
these cells with different HIV-1 or HIV-2 isolates. Even with ahigh multiplicity
ofHIVs,
everyattempt
met withfailure.
Transcription
directedby
the HIVLTRs was showntobe frans-activatedby superinfection
withherpes
simplex
virustype
1(HSV-1).20
F12 clone cells were thus infected with1, 4,
or 10plaque-forming
unitsJLL
JLUL 1XL
^p ^^^^. ^^^^_
FIG. 1.
Radioimmunoprecipitation
of[35S]cysteine-labeled
cellularlysates
of HTLV-IIIB-infected HUT-78 cells(c
+)
and D10 and F12clones. About4 x 106cpm per conditionwasincubated with humannegative
(
—
)
orHIV-posi-tive
(
+)
serumandprocessed
asdescribed. Thearrowsindicatethemajor
viralantigens
detectable in the RIPAtest.gp160*
gp120»
p55* gp4l» p24*p19
p17
389
(PFU)
percellofHSV-1,
butnoHIVproduction
wasdetectable underanyconditions,
evenifthesuperin-fecting
HSV-1 didreplicate
in F12 as well as in HUT-78 control cells(data
notshown).
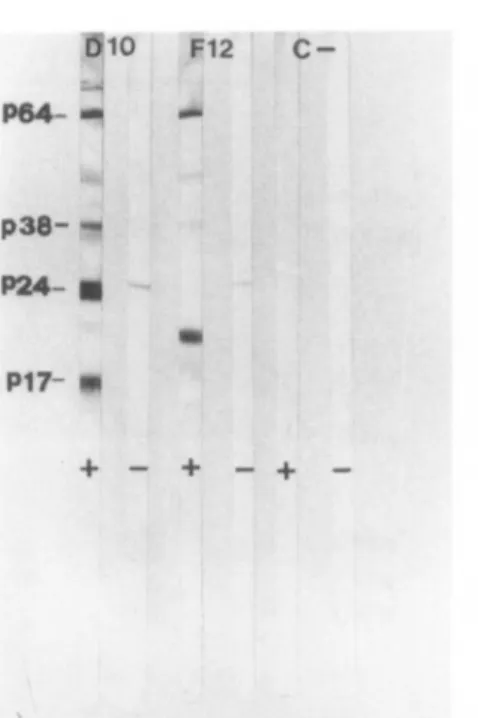
Proteinpattern
analysis
The
productively
infectedDIO clone shows a viralprotein
pattern
resembling
that ofHUT-78/HTLV-HIB cells
except
for the lower band of thep24-25
doublet and theelectrophoretic mobility
ofgpl20,
possibly
dueto adifferentpattern
ofglycosylation (Fig.
1).
In contrast, in the F12 cell clone there
is
a consistent reduction inp55
gag precursor, which ispoorly
cleaved with the
consequent
almost total absence ofp24
andpl7
gagproteins.
Further,
only
gpl60
pre-cursor is
present
but there is a lack ofgpl20
andgp41, easily
visible in HUT-78/HTLV-IIIB and in DIOcells. Pulse-chase
experiments
didnotshowanycleavage
ofgpl60
in F12 clone cellseven9 hours after theremoval of radioactive label
(data
notshown).
In DIO and F12
cells,
p64
andp38 proteins
(the
reversetranscriptase
and the viralendonuclease,
respec-tively)
are visibleby
WBanalysis
whilebeing
undetectableby
RIPA. In F12 cells there is aprominent
band ofa
protein
withanMr
of 19kDa,
also visibleby
WBanalysis (Fig.
2),
thatmaybe eitheranaberrantproduct
of thegag geneortheproduct
of therev HIV exone. Todefine moreexactly
the natureof thisprotein species,
we have labeled HTLV-IIIB-infected HUT-78 and F12 cells with[32P]orthophosphoric
acid
(Fig.
3).
The failuretoreveal substantial differencesin the RIPA of32P-labeled
proteins
between thepositive
control and the F12clone,
thatis,
theabsence of any 19 kDasignal,
suggests
that thisprotein
does notbelong
tothephosphorylated
gagantigen family. Conversely,
thespecies
mostrepresented
is thep27
nef
protein,
which ispresent
atcomparable
levels in both F12 andHTLV-IIIB-infected HUT-78 cells. Thepossibility
ofa double infectionby
HTLV-I and HIV-1 in the F12 clone(a
pl9 protein
is a coreprotein
ofHTLV-I)
has been ruledoutby
(1)
utilizing cytoplasmic
IFAwith monoclonal antibodiesagainst
the HTLV-Ipi9
coreprotein,
and(2)
hybridizing
F12DNA withan HTLV-Iprobe
of fullgenomic length
(data
notshown).
F12
P38-.«
P24- g P17- •• + - + -+-FIG. 2. Westernblot
analysis
of uninfected HUT-78(C
-), DIO,and F12 cell
lysates.
The nitrocellulosestrips
wereincubated with human
negative
(—
)or
HIV-positive
(+)
serum. On the left are indicated theposition
of themoreFIG. 3.
Radioimmunoprecipitation
of[32P]orthophosphoric
acid-labeled cellularlysates
of HTLV-IIIB-infected HUT-78cells(c+)
and F12 clone. About3 x 105(lanesAandB),
5 x 105(lanes
C andD),or106(lanes
F, G,andI)
cpm per condition was incubated with an
HIV-positive
(
+) ornegative
(lanesE and H, 5 x 105cpm)
serum andprocessed
as described. On the leftare indicated themajor
viralantigens
detectable in the RIPA test(the
p55
gagprecursorand the
p27
nef
protein);
ontheright
are indicated theMT
of the standards.DAM
analysis
The
pattern
analysis (Fig.
4)
ofHIVproviral
DNA restrictedby
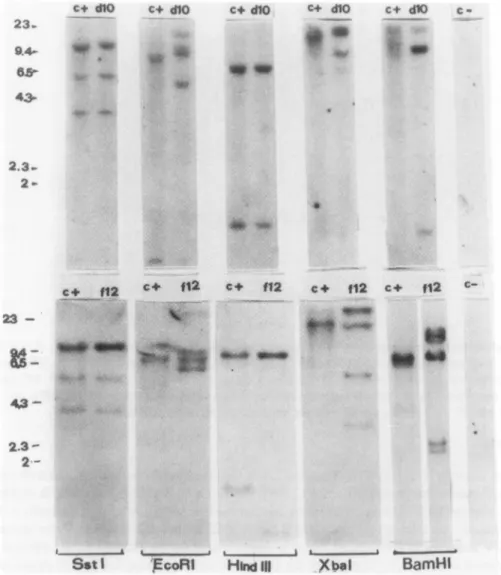
the SstI enzyme, which cleaves in the LTRregions,13
shows that both DIO and F12 clones have an HIV genome ofalength
similar tothat ofHTLV-IIIB-infected HUT-78
cells,
thusruling
outthat the lack of HIV release in F12cellsis duetomajor
genomic
deletions. Asalready
demonstrated for HTLV-IIIB-infected H9cells,13
the presence of twocoexisting
HIV genomesinDIO and F12 cells is demonstratedby
the SstI and EcoRIpatterns.
Most restriction enzymes
(six
ofeight
forthe DIOcloneand six ofnine for the F12clone,
notall shown inFig.
4)
generate
differentcleavage
patterns
withrespect
to HUT-78/HTLV-IIIB control cells. Incon-trast, the
heterogeneity
is lesspronounced
between thetwoclones(e.g.,
EcoRI)
andbetween each clone and the infectedparenteral
cellpopulation,
from whichthey
derive(data
notshown).
The Xbal enzyme does notrecognize proviral
sites in HTLV-IIIB-infected HUT-78 and in DIO cells. Thattwo additional bandsofhigh
molecularweight
aregenerated only
in F12 clone may beinterpreted
asthe appearance ofanXbal-specific
internal site inoneof thetwocoexisting
HIV genomes.Finally,
asreported
for the H9/HTLV-IIIBcells,3,10
the restrictionpattern
of the two clones did notchange noticeably
even after 12months of culture(data
notshown).
RNA
analysis
Several studies of
HIV-specific
RNAs21,22
reported
the presence in HIV-infected cells of threemajor
bandsat9.3, 4.3,
and 1.8-2kb,
representing, respectively,
thegenomic length transcript,
theenv-specific
mRNA,
andafamily
of minormessengerscoding
fortheregulatory
HIVproteins.
An additionaltranscript
was
occasionally
detectedat5kb.23
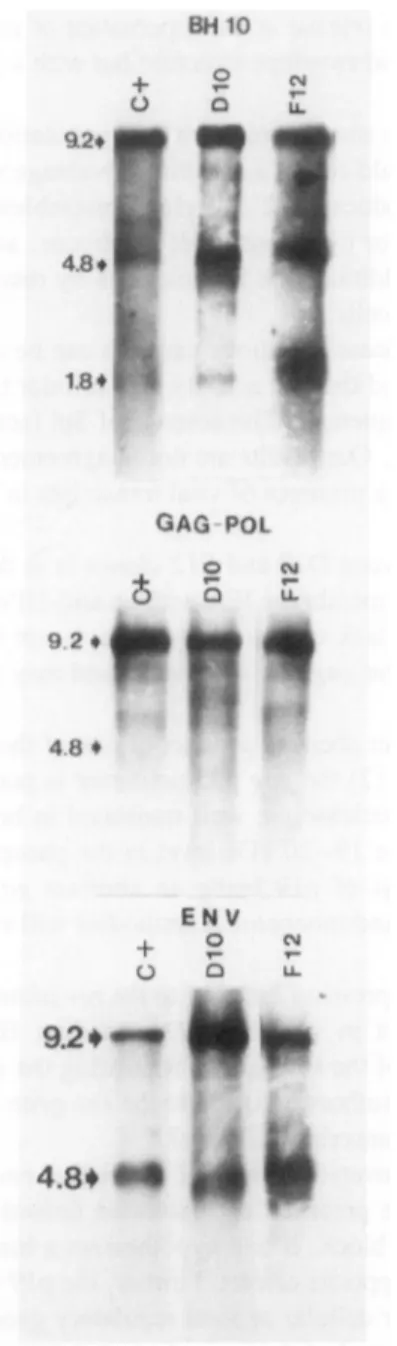
The
hybridization
ofintracellular viralpoly-A
+ RNAwith thefull-length genomic
BH10probe
reveals the same threemajor
bands in bothpositive
controlcells,
inDIO,
and F12 clones(Fig.
5).
The evidentoverproduction
of 1.8-2 kb viral RNA in the F12 clonehybridized
withaBH10probe
canhypothetically
fit with the
overexpression
ofa 19 kbregulatory
protein
in the F12 clone(Fig.
1).
Hybridization
ofthe sameRNAs with thegag-pol
or envsubgenomic probes
fails to show anyquantitative
(as
shown alsoby
c+ dtO c+ dlO c-f dtO c+ <flo c+ dlO c• .; : 4Êt *fr c+
fl2
c+ «2 c+ ft2 c*(12
<3- 2.3-2SstI
pcoRl
Hind
III XbalBamHI
FIG. 4. Restriction enzyme
analysis
ofHIV DNAintegrated
inDIOand F12 genomes. Eachpattern wascompared
with that of HTLV-IIIB-infected HUT-78 cells
(c
+);(c-),
uninfected HUT-78 cells. At the bottomareindicated the restriction enzymesemployed;
onthe leftare indicatedthe kilobasepairs (kbp)
of the \ DNA markercutby
Hindlll.hybridization
with theglucose-6'-phosphodehydrogenase
probe)
orqualitative
differences amongDlO,
F12,
and HUT-78/HTLV-IIIB cells(Fig.
5).
DISCUSSION
The
cloning
ofHUT-78 cellsinfected withanRT-positive
supernatant
of PBLfromanAIDSpatient
ledtotheisolation oftwoclones
harboring
differentHIVvariants,
one(DlO)
persistently
infectedby
areplica-tion-competent
HIV,
the other(F12)
exhibiting
anintegrated
HIV genome unable togenerate
infectious viralparticles.
The isolation of such clonesemphasizes
thepossibility
ofrecovering
more than one HIVvariant from PBL of the same AIDS
patient.
Moreinterestingly,
fromthe same virus isolation assay wehave detected adefectiveHIV variant
that,
onceintegrated
in the host genome, is able toprotect
the cells from HIVsuperinfection through
aphenomenon
ofhomologous
viral interference.The simultaneous presence of more than one variant in cell
populations
infected with HIV was firstreported by Wong-Staal
etal.,3
whodescribedthe coexistence ofatleasttwodifferentgenotypes
in 2 of18 AIDS isolates. This evidencecamefromrestrictionpattern
analysis
ofintegrated
viralDNAusing
enzymesBH10 o a u. 9.2* 4.8» 1.8*r 92» GAG POL o m o
S
£ M*^^ ^^^^ JIB9.2*
4.8*
ENV i O CM ' T- T-Ü Q LL ^^^m Hft ^^^m.-kit
FIG. 5. Northern blot
analysis
ofviral RNA: 3-5(Jigpoly-A+
RNAofDIO, F12,andHTLV-IIIB-infectedHUT-78 (c+)cellswerehybridized
withthenick-translated fullgenomic length
BH10probe,
withsubgenomic gag-pol
or envprobes,
bothrepresenting
themostconserved sequences oftherespective
exones. Thearrows ontheleft indicate thekbp
of theHIV RNAmajor signals.
The
possibility
thatproviral
genomespresent
in DlOand F12 clonesoriginated
fromin vitro mutations cannot be ruled out, sincesomany culture passages werenecessary toestablish theclones.However,
thishypothesis
isstrongly
in contrast to the in vitrogreat
genomic stability
ofintegrated proviral
HIV,
asalready
showncomparing proviral
restrictionpatterns
after3,10 9,3
or even 12months(our
results forDlOand F12
clones)
of culture.The
nonproducer
HIV-infectedF12 clone may haveoriginated
fromanHIV-defectiveparticle originally
present
in the AIDSpatient
and released from thepatient's
PBL. This virus could have infected CD4+HUT-78
cells,
taking
advantage
of the presence ofacoinfecting replication-competent
HIV. Such viralcomplementation
could inturn leadto release in thesupernatant
of reinfected fresh HUT-78 cells of viralparticles
with normalnucleocapside
andenvelope
structurebut withagenome unabletocode for infectious HIV virions.Both DlO- and F12-infected clones may
represent
a cellpopulation originally harboring
HIV variantswith limited
cytopathic
effects that could conferaselectiveadvantage
withrespect
tocellsinfectedby
morecytopathic
HIV variants. Thenonproducer
F12 cell clone resembles the DlO clone in terms of cellulargrowth,
percentage
of cellspositive
forcytoplasmic
HIVantigens,
and downregulation
of CD4receptor
sites and viralRNAtranscripts.
Inaddition,
the F12 clone isby
restrictionpattern
much more related to DlO thantothe HUT-78/HTLV-IHB cells.The
inability
of the F12clone torelease infectiousparticles
canbe duetoboth viral and cellularfactors.Among
thelatter,
Jonesetal.24
showed that theactivity
ofacellulartranscription
factor(Spl)
isnecessary for thecorrecttranscription
ofHIV sequences. The absenceofSpl
factor leadstoa 10-foldreductioninthein vitroHIV
transcriptional efficiency.
Our resultsarenotinagreement
withthehypothesis
ofaSpl
defectin F12clone becauseofthe
comparable
presence of viraltranscripts
inDIO,
Fl2,
and HTLV-IIIB-infected HUT-78 cells(Fig.
5).
The most
important
difference between DlOandF12 clones is atthe level of viralprotein
pattern.
The very lowpositivity
of F12cellsby
themembrane IFA withananti-HIV antiserum fits well with the absenceof
cleavage products
ofgpl60.
The lack ofgpl60 cleavage
is not due to a defect in theviral-specific
protease,
whichplays
aroleonly
in thegagp55
cleavage25
and may beduetoa mutation inthecleavage
site.
F12
pl9
canhardly
be considered an aberrantproduct
ofoneofthe three structural genes, since(1)
theenv
gpl60
issubstantially
uncleaved,
(2)
thegagp55
precursor ispoorly expressed,
and(3)
thep64
viralpolymerase
as well as thep34
endonuclease are well translated in both F12 and DlO clones(Fig.
2).
Inaddition,
the absence ofasignal
atthe 19-20kDa level inthephosphorylated protein
pattern
of the F12 clone does notsupport
thehypothesis
ofpl9 being
an aberrant gagproduct (Fig.
3).
Experiments
inprogress
utilizing
anti-revpolyclonal
andmonoclonalantibodieswill establish whether theF12pl9 protein
is theproduct
of therevgene.Sodroskiet
al.26
assigned
ananíí-repression
functiontotherevprotein
to counterthe effects ofcw-acting
negative regulatory
sequencespresent
in viral mRNAsencoding
HIV structuralproteins. Conversely,
Feinberg
etal.22
hypothesized
arole of therevgene inregulating
thesplicing
of viral mRNAs.Regardless
of the mechanism of
action,
allthese authorsassigned
totherevgene apositive regulatory
function ofgagand
pol
protein synthesis
attheposttranscriptional
level.It is thus difficult to correlate the
overexpression
ofaputative
revpl9 protein
with reduced gagp55
production
unless wehypothesize
the presence of a mutated defective rev.Alternatively,
the increasedpresence of
pl9
may accountfor thisblock,
ifonehypothesizes
abimodal effect ofrevprotein by
whichlow and
high
amountswould induceopposite
effects.Further,
the p 19overproduction
may bearesponsetosomeundefined block relatedtoeither cellularorviral
regulatory
genes.Anaccurate
study
of the sequences and of thesecondary
structureofviral mRNA willalso be useful toexactly
understand both themechanism(s)
of translation inhibition of gagspecific
messengers and theorigin
oftheuncleavage
ofenvgpl60.
ACKNOWLEDGMENTS
This workwas
supported
inpart
by
grants
from AIDSProject
1987-1988 oftheMinistry
ofHealth,
Rome,
fromGruppo
diVirologia
87.01640.04Consiglio
Nazionale delleRicerche, Rome,
and from the Associazione Italianaperle Ricerche sul Cancro.WeareindebtedtoMs. A. Guderzofor excellent secretarial assistance.
REFERENCES
1. Rossi
GB,
VeraniP, MacchiB,FedericoM,OrecchiaA,NicolettiL,ButtóS,LazzarinA,MarianiG,Ippolito
G,
and ManzariV:
Recovery
of HIV-related retroviruses from italianpatients
with AIDS orAIDS-relatedcomplex
2. Evans
LA,
McHugh
TM,
StitesDP,
andLevy
JA: Differentialability
ofhumanimmunodeficiency
virus isolatestoproductively
infecthumancells. JImmunol1987;138:3415-3418.
3.
Wong-Staal
F, ShawGM, Hahn BH, Salahuddin SZ,Popovic
M, Markham P, RedfieldR, and Gallo RC: Ge-nomicdiversity
of humanT-Lymphotropic
virustypeIII(HTLV-III). Science 1985;2:759-762.4. HahnBH,ShawGM,
Taylor
ME, RedfieldRR, Markham PD, Salahuddin SZ,Wong-Staal
F, GalloRC,
Parks ES, and Parks WP: Genetic variation in HTLV-III/LAV overtime inpatients
with AIDS or atrisk for AIDS. Science1986;232:1548-1553.
5.
Magasiny
S,Spire
B, Barré-SinoussiF, and Chermann JC: Genomicvariability
of selected LAV-related AIDS retroviruses. AIDS Res1986;2:19-30.
6. Devare SG, Srinivasan A, Bohan CA,
Spira
TJ, CurranJW, andKalyanaraman
VS: Genomicdiversity
of theacquired immunodeficiency
syndrome
retroviruses is reflected in alteration of its translationalproducts.
Proc Nati Acad Sei USA1986;83:5718-5722.
7. Benn S,
Rutledge
R, Folks T, Gold J, BakerL, McCormick J, Feorino P, Piot P,Quinn
T, and Martin M: Genomicheterogeneity
of AIDS retroviral isolates from North America and Zaire. Science 1985;230:949-951. 8. SrinivasanA,YorkD,Ranganathan
P,Ferguson
R,ButlerDJr,FeorinoP,Kalyanaraman
V,JaffeH,CurranJ,and Anand R: Transfusion-associated AIDS:
Donor-recipient
humanimmunodeficiency
virus exhibitsgenetic
het-erogeneity.
Blood1987;69:1766-1770.
9. Starcich BR, HahnBH, ShawGM,
McNeely
PD, ModrowS, WolfH, ParksES, Parks WP,Josephs
SF,
Gallo RC,andWong-Staal
F: Identification and characterization of conserved and variableregions
in theenvelope
gene ofHTLV-III/LAV,the retrovirus of AIDS. Cell 1986;45:637-648.10. HahnBH,GondaMA,ShawGM,
Popovic
M,HoxieJA,GalloRC,andWong-Staal
F: Genomicdiversity
of theacquired
immunedeficiency syndrome
virus HTLV-III: Different viruses exhibitgreatestdivergence
in theirenve-lope
genes. Proc Nati Acad Sei USA1985;82:4813-4817.
11. Clements JE,Pederson
FS,
Narayan
0, and Haseltine WA: Genomicchanges
associated withantigenic
variation ofVisna vimsduring
persistent
infection. Proc Nati Acad Sei USA 1980;77:4454.12. Montelaro RC, Parekh B,
Orrego
A, and Issel CJ:Antigenic
variationduring persistent
infectionby equine
infectious anemiavirus,a retrovirus.JBiol Chem 1984;259:10539.13. HahnBH,ShawGM,
Arya
SK,Popovic
M,GalloRC,andWong-Staal
F: Molecularcloning
and characterization of the HTLV-m virus associated with AIDS. Nature1984;312:166-169.
14.
Koyanagi
Y,MilesS,
Mitsuyasu
RT,MerrillJE,VintersHV,and Chen ISY: Dual infection of the centralnervoussystem
by
AIDS viruses with distinct cellulartropisms.
Science 1987;236:819-821.15. Von Bliesen H, BeckerWB, Heneo K, Helm EB, Gelderblom HR, Brede HD, and
Rubsamen-Waigmann
H: Isolationfrequency
andgrowth properties
of HIV-variants:Multiple
simultaneous variants in apatient
demon-strated
by
molecularcloning.
J Med Virol 1987;23:51-66.16. Robert-GuroffM,Ruscetti
FW,
PosnerLE,
PoieszBJ, and Gallo RC: Detection of the human T-celllymphoma
viruspl9
in cells ofsomepatients
withcutaneousT-celllymphoma
and leukemiausing
amonoclonalantibody.
JExpMed
1981;154:1957-1964.17.
Gathings
WE, Lawton AR, andCooper
MD: Immunofluorescent studies of thedevelopment
ofpre-B
cells, Blymphocytes
andimmunoglobulin
isotype
diversity
in humans. Eur JImmunol 1977;7:804.18.
Chirgwin
JM,Przybyla
AE,MacDonaldRJ,and Rutter WJ: Isolation ofbiologically
active ribonucleic acid fromsourcesenriched in ribonuclease.
Biochemistry
1979;18:5294-5299.19. HoxieJA,
Alpers
JD,RackowskiJL, HuebnerK,Haggarty
BS,CedarbaumAJ,
and Reed JC: Alterations in T4(CD4)
protein
and mRNAsynthesis
in cells infected with HIV. Science1986;234:1123-1127.
20. MoscaJD,BednarikDP,
Raj
NBK, RosenCA,SodroskiJG, HaseltineWA,and Pitha PM:Herpes simplex
virustype-1
canreactivatetranscription
of latent humanimmunodeficiency
virus. Nature1987;325:67-70.21.
Muesing
MA, Smith DH, Cabradilla CD, Benton CV,Lasky
LA, andCapon
DJ: Nucleic acid structure andexpression
of the humanAIDS/lymphadenopathy
retrovirus. Nature1985;313:450-458.
22.
Feinberg
MB,JarrettRF, AldoviniA,GalloRC,
andWong-Staal
F: HTLV-IIIexpression
andproduction
involvecomplex regulation
atthe levels ofsplicing
and translation of viral RNA. Cell1986;46:807-817.
23. Folks TM, Powell D,
Lightfoote
M,Koenig
S, Fauci AS, Benn S, RabsonA,Daugherty
D, Gendelman HE,Hoggan
MD, VenkatesaS,
and Martin MA:Biological
and biochemical characterization ofacloned leu-3~ cellsurviving
infection with theacquired
immunedeficiency syndrome
retrovirus. JExp
Med1986;164:280-290.
24. Jones
KA,
Kadonaga
JT, Luciw PA, andTjian
R: Activation of the AIDS retrovirus promoterby
the cellulartranscription
factorSpl.
Science1986;232:755-758.
25. Kramer
RA,
Schaber MD, Skalka AM,Ganguly
K,Wong-Staal
F,
andReddy
EP: HTLV-III gagprotein
isprocessed
inyeastcellsby
the viruspo/-protease.
Science1986;231:1580-1584.26. SodroskiJ, GohWC, RosenC,
Dayton
A,Terwilliger
E, HaseltineW: A secondpost-transcriptional
trans-acti-vatorgene
required
for HTLV-IIIreplication.
Nature 1986;321:412-417.Address
reprint
requests
to: Giovanni B. RossiLaboratory of
Virology
Istituto
Superiore
di Sanità V.leRegina
Elena,
299 00161 RomeItaly
1. Andrey Skripchenko, Stephen J. Wagner, Dedeene Thompson-Montgomery, Helen Awatefe. 2006. Thiazole orange, a DNA-binding photosensitizer with flexible structure, can inactivate pathogens in red blood cell suspensions while maintaining red cell storage properties. Transfusion 46:2, 213-219. [CrossRef]
2. Stephen J. Wagner, Andrey Skripchenko, Louis Cincotta, Dedeene Thompson-Montgomery, Helen Awatefe. 2005. Use of a flexible thiopyrylium photosensitizer and competitive inhibitor for pathogen reduction of viruses and bacteria with retention of red cell storage properties. Transfusion 45:5, 752-760. [CrossRef]
3. M. Federico, F. Nappi, R. Bona, P. D'Aloja, P. Verani, G.B. Rossi. 1995. Full expression of transfected nonproducer interfering HIV-1 proviral DNAabrogates susceptibility of human He-La CD4+ cells to HIV. Virology 206:1, 76-84. [CrossRef]
4. P. Verani, S. Buttò, B. Taddeo, M. Federico, G.B. Rossi. 1993. HIV variability and perspectives for a vaccine. Vaccine
11:5, 542-544. [CrossRef]
5. A.M. Luciani, A. Rosi, M.T. Maggiorella, M. Federico, N. Sulli, P. Verani, G.B. Rossi, V. Viti, L. Guidoni. 1991. Interaction of HIV-1 with susceptible lymphoblastoid cells 1H NMR studies. FEBS Letters 285:1, 11-16. [CrossRef]

![FIG. 1. Radioimmunoprecipitation of [35S]cysteine-labeled cellular lysates of HTLV-IIIB-infected HUT-78 cells](https://thumb-eu.123doks.com/thumbv2/123dokorg/7578907.112367/5.876.278.521.632.981/radioimmunoprecipitation-cysteine-labeled-cellular-lysates-htlv-iiib-infected.webp)
![FIG. 3. Radioimmunoprecipitation of [32P]orthophosphoric acid-labeled cellular lysates of HTLV-IIIB-infected HUT-78 cells (c + ) and F12 clone](https://thumb-eu.123doks.com/thumbv2/123dokorg/7578907.112367/7.876.157.662.112.441/radioimmunoprecipitation-orthophosphoric-labeled-cellular-lysates-htlv-iiib-infected.webp)