Research Article
Mapping QTL for Root and Shoot Morphological
Traits in a Durum Wheat
×T. dicoccum Segregating
Population at Seedling Stage
Anna Iannucci,
1Daniela Marone,
1Maria Anna Russo,
1Pasquale De Vita,
1Vito Miullo,
1Pina Ferragonio,
1Antonio Blanco,
2Agata Gadaleta,
2and Anna Maria Mastrangelo
11Consiglio per la Ricerca in Agricoltura e l’analisi dell’economia Agraria-Centro Cerealicoltura e Colture Industriali (CREA-CI),
SS 673 km 25.2, 71122 Foggia, Italy
2Department of Soil, Plant and Food Sciences, Section of Genetic and Plant Breeding, University of Bari Aldo Moro, Via G. Amendola
165/A, 70126 Bari, Italy
Correspondence should be addressed to Anna Maria Mastrangelo; [email protected] Received 12 January 2017; Revised 12 May 2017; Accepted 21 June 2017; Published 6 August 2017 Academic Editor: Mohamed Salem
Copyright © 2017 Anna Iannucci et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
A segregating population of 136 recombinant inbred lines derived from a cross between the durum wheat cv.“Simeto” and
the T. dicoccum accession “Molise Colli” was grown in soil and evaluated for a number of shoot and root morphological
traits. A total of 17 quantitative trait loci (QTL) were identified for shoot dry weight, number of culms, and plant height
and for root dry weight, volume, length, surface area, and number of forks and tips, on chromosomes 1B, 2A, 3A, 4B, 5B, 6A, 6B, and 7B. LODs were 2.1 to 21.6, with percent of explained phenotypic variability between 0.07 and 52. Three QTL were mapped to chromosome 4B, one of which corresponds to the Rht-B1 locus and has a large impact on both shoot
and root traits (LOD 21.6). Other QTL that have specific effects on root morphological traits were also identified.
Moreover, meta-QTL analysis was performed to compare the QTL identified in the “Simeto” × “Molise Colli” segregating
population with those described in previous studies in wheat, with three novel QTL defined. Due to the complexity of
phenotyping for root traits, further studies will be helpful to validate these regions as targets for breeding programs for
optimization of root function for field performance.
1. Introduction
The root system architecture defines the shape and spatial arrangements of the root structure within the soil [1]. A number of factors contribute to the definition of the mor-phology of the root system, such as the angle and rate of root growth, and the diameters of the individual roots. The devel-opment of the root structure depends on interactions between the genetic features of a plant and the environment in which the roots grow (i.e., soil type and composition, water
and nutrient availability, and microorganism profile). The
main challenge in studying root traits is the need for robust and high-throughput methods for phenotypic evaluation
that can provide a proxy for field performance, because the
measurement of root traits under open field conditions
can be very difficult. This is particularly the case for genetic studies that require analysis of large sets of samples. For this reason, various hydroponic culture techniques have been adopted, together with experimental systems with soil-based growth substrates that can offer better tools to predict
plant behavior under field conditions, for example [2–5].
Once young seedlings have been grown in the laboratory or in glasshouses with various methods [6, 7], scanner-based image analysis can make root analysis less time consuming. Therefore, although there is the limitation of the early growth stage of the plants analyzed, this represents a
Volume 2017, Article ID 6876393, 17 pages https://doi.org/10.1155/2017/6876393
decisive tool, as it allows the evaluation of large sets of genotypes, as in the case of segregating populations or association-mapping panels.
Many studies have been carried out to dissect out the genetic basis of root system architectures. For cereals, quan-titative trait loci (QTL) for root traits have been mapped in
rice (Oryza sativa L.) [8–14], maize (Zea mays L.) [15–17],
sorghum [18], and bread and durum wheat [4, 5, 19–37].
Linkage mapping has largely been used to map QTL for root traits in biparental populations, although recently, associa-tion mapping with germplasm collecassocia-tions has also been used in durum wheat [35, 37]. Altogether, these studies have shown relatively complex genetic control for root traits and strong environmental effects, with only a few examples of QTL that can individually explain up to 30% of the pheno-typic variation in rice [38, 39] and maize [40] and up to 50% in wheat [4]. In most cases, root traits are regulated by
a suite of small-effect loci [41]. Uptake of water and nutrients,
anchorage in the soil, and interactions with microorganisms are among the main functions of the root structure, and these are responsible for crop performance, in terms of yield and quality. Indeed, some studies have shown overlap of QTL for root features with QTL for traits related to productivity [35, 37, 42–45]. Moreover, despite the complexity of the genetic control of root traits, there are some examples in which marker-assisted selection for root QTL has been suc-cessfully exploited to improve the root-system architecture and yield in rice [45, 46] and maize [40, 47].
Grain yield in wheat has been greatly improved over the last century, with the introduction of semidwarf wheat culti-vars that are characterized by higher harvest index compared to taller genotypes [48]. Many studies have focused on the relationships between above-ground biomass and root growth; nevertheless, controversial results have been reported. In general, recent studies have indicated the absence of a clear correlation between shoot and root growth in wheat, with shoot and root traits reported to be controlled
by different sets of genetic loci [21, 49–51]. Different
conclu-sions have been reported in other studies. Miralles et al. [52]
indicated that spring wheat plants with dwarfing genes are
characterized by reduced plant height but increased root length and dry weight. More recently, Kabir et al. [36] ana-lyzed two bread-wheat segregating populations and defined a negative correlation between root traits and plant height in both of the populations. On the contrary, Subira et al. [53] indicated that the Rht-B1b dwarfing allele is effective in reducing both aerial and root biomass in durum wheat.
In the present study, a durum wheat (Triticum turgidum L. var. durum) population of 136 recombinant inbred lines (RILs) was grown in soil under controlled conditions to identify the chromosome regions that are involved in the control of their root and shoot architecture.
2. Materials and Methods
2.1. Genetic Materials. The RIL population of 136 F6 lines that was used in the present study was developed from a cross
between the Italian durum wheat cv. “Simeto” (Capeiti/
Valnova) and a cultivar of T. dicoccum known as“Molise Colli”
that was selected within the framework of a local population of T. dicoccum (from the Regional Agency for Development and Innovation of Agriculture of the Molise Region (Agenzia Regionale per lo Sviluppo e l’Innovazione dell’Agricoltura della Regione Molise)).
2.2. Plant Growth and Soil Sampling. The RIL population and the parents were grown in plastic cylinders containing a soil mixture (soil: sand, 50 : 50; v/v). Before the pot experiments, soil with a history of exposure to annual cereal species was collected (in July 2013) from the experimental farming
sta-tion of the Cereal Research Centre in Foggia (Italy; 41°28′
N, 15°34′ E; 76 m a.s.l.). The samples were collected from
the upper 30 cm of the soil profile and air dried for 1
week. They were then thoroughly mixed, passed through a 2 mm sieve (to remove gravel fragments), cleaned of
plant debris, and stored in a cold room (4°C) until further
use. This soil was an unsterilized loam soil (USDA classi
fica-tion system) with the following characteristics: 21% clay, 43%
silt, 36% sand, pH 8 (in H2O), 15 mg/kg available
phospho-rous (Olsen method), 800 mg/kg exchangeable potassium
(NH4Ac), and 21 g/kg organic matter (Walkey-Black
method). Silica sand with a grain size that ranged from 0.4 mm to 0.1 mm was used. The soil mixture is hereinafter
referred to as“soil.”
Before sowing the seeds, they were surface sterilized by soaking them in 2% sodium hypochlorite for 5 min and then rinsed several times with distilled water. The seeds were put
into Petri dishes with one sheet of filter paper (Whatman
number 1) that was moistened with 5 mL distilled water, and these were kept in a dark incubator at a constant temper-ature of 20°C for 48 h. Three germinated wheat seeds (roots, <1 cm) of each genotype were seeded into each of the plastic pots (diameter, 7 cm; height, 26 cm) that contained 1.3 kg
soil, and then 40 kg/ha NH4NO3 (26% elemental nitrogen)
was applied. The pots were lined with a filter paper
(Whatman 3MM) to avoid soil loss. Immediately after sowing, 200 mL deionized water was added to each pot. To maintain the soil moisture, the seedlings were regularly
watered at 3-day intervals to 70% offield capacity. The pots
were placed in a growth chamber with a 16 h/8 h light/dark
period at 20°C/16°C, with a light intensity of 1000μmol
photons/m2/s photosynthetically active radiation at the leaf
surface. The experiments were performed using a completely randomized design, with four replicates.
After emergence, the seedlings were thinned to one plant per pot. These plants were grown until they were at the 5th leaf developmental stage (Zadoks growth scale 15; [54]). When they reached this stage, the days after sowing, maxi-mum shoot lengths (cm), and number of shoots were recorded. The plants were collected by pulling them from the soil in the pots, with all of the plant material manually removed from the pot and the shoots and roots washed with
deionized water. The roots were stored at 4°C in 75% ethanol,
to preserve the tissue until all of the analyses had been done. After the analysis, the aerial parts of the samples and the
roots were oven dried for 72 h at 70°C andfinally weighed,
to obtain the shoot dry weight (mg/plant) and root dry weight (mg/plant).
2.3. Scanner-Based Image Analysis. The root measurements were performed using the Win-RHIZO system (version 4.0b; Regent Instruments Inc., Quebec, Canada), which is an interactive scanner-based image analysis system for scanning, digitizing, and analyzing of root samples. A Windows-based PC, with a Pentium (R) D CPU and 992 MB RAM, and a scanner (Perfection V700/V750 2.80A; Epson) set to a scanning resolution of 200 dots per inch (dpi: 118.11 dots per cm) were used. The scanner had two light sources, one located above, on the scanner cover, and the other below, incorporated in the main body of the scan-ner. The root samples were placed in a plexiglass tray (20 cm × 30 cm) with a 4 mm to 5 mm deep layer of water. They were adjusted to help untangle the roots and to mini-mize root overlap. Several morphological traits of the roots were recorded using the root analysis software, as length
(cm), surface area (cm2), mean diameter (mm), volume
(cm3), and number of tips, forks, and crossings. All of these
parameters were measured individually, and for length, sur-face area, volume, and tips, the roots were then classified into 10 different classes based on their diameters: class 1, 0.0– 0.5 mm; class 2, 0.5–1.0 mm; class 3, 1.0–1.5 mm; class 4, 1.5–2.0 mm; class 5, 2.0–2.5 mm; class 6, 2.5–3.0 mm; class
7, 3.0–3.5 mm; class 8, 3.5–4.0 mm; class 9, 4.0–4.5 mm; and
class 10, >4.5 mm.
2.4. Statistical Analysis. The means, standard deviations,
coefficients of variation, and ranges of each measured
mor-phological trait were separately calculated for each genotype. The data were also analyzed using ANOVA, and the homo-geneity of the phenotypic variance between the replications was verified, with the means separated by Fischer’s protected least-significant difference at P < 0 05 for all of the traits, to test the differences across the RILs and the two parents
(i.e.,“Molise Colli” and “Simeto”). The heritability (H) was
estimated for each trait, as in
H =σb2
σt2
, 1
where σb2 and σt2 are the between-line variance (as an
estimate of the genotypic variance) and the total variance (as an estimate of the phenotypic variance), respectively, as estimated from the mean squares of the analysis of variance. To obtain a general comprehensive characterization of the samples, the root traits were subjected to principal com-ponent analysis (PCA) based on correlations, followed by factor analysis. To overcome differences in size during recording, the data for the different traits were standardized to a mean of zero and variance of one [55]. The components that represented the original variables (traits) were also
extracted. Only those with eigenvalues≥ 3.0 were considered
as having a major contribution to the total variation [56]. The first and second principal component axis scores were
plotted to aid the visualization of the genotype differences.
All of the statistical analyses were performed using the JMP software (version 8.0; SAS Institute Inc.).
Genome-wide QTL searches were conducted on the “Simeto” × “Molise Colli” linkage map that was described
by Russo et al. [57]. Briefly, the map covers all of the 14 chromosomes of the durum wheat genome with 9040
markers on 2879.3 cM. The“Simeto” × “Molise Colli”
segre-gating population (SMC) was also genotyped with the markers BF-MR1 and BF-WR1, to infer the presence of the alleles Rht-B1b and Rht-B1a, respectively [58]. The inclusive composite interval mapping method [59] was used for the QTL mapping, with the QGene 4.0 software [60]. The scanning interval of 2 cM between markers and putative QTL with a window size of 10 cM were used to detect QTL. The marker cofactors for background control were set by single marker regression and simple interval analysis,
with a maximum of five controlling markers. Putative QTL
were defined as two or more linked markers that were
associ-ated with a trait at a log10odds ratio (LOD)≥ 3. Suggestive
QTL at the subthreshold of 2.0 < LOD < 3.0 are reported
for further investigations [61]. For the main QTL effects,
the positive and negative signs of the estimates indicated
that “Simeto” and “Molise Colli,” respectively, contributed
toward higher value alleles for the traits. The proportion of phenotypic variance explained by a single QTL was determined by the square of the partial correlation coefficient
(R2). Finally, the 95% confidence intervals of the QTL were
estimated using the approach of Darvasi and Soller [62], as given in
Confidence interval = 163
N × R2, 2
whereN is the population size and R2is the proportion of the
phenotypic variance explained by the QTL.
2.5. Meta-QTL Analysis. Twelve previously published studies
were identified that reported QTL for root traits (see
Supple-mentary Table S1 available online at https://doi.org/10.1155/ 2017/6876393). An integrated map of the A and B genomes was constructed using the wheat map developed with a 90K single-nucleotide polymorphism (SNP) array [63] on which a durum-wheat linkage map was projected, based on the SNP and simple sequence repeat (SSR) markers (“Cic-cio” × “Svevo”) by Colasuonno et al. [64], together with the SMC genetic map. The map obtained was used as the refer-ence map for merging another wheat consensus map that included diversity array technology (DArT) and PCR-based markers, as described in Marone et al. [65, 66]. The chromo-somal regions that contained QTL for root traits retrieved in the literature were also integrated. All of the calculations for both the creation of the integrated map and the QTL projec-tions were performed with the Biomercator software v.4. For n individual QTL, the Biomercator software tests the most likely assumption between 1, 2, 3, 4, and n QTL. The Akaike information criterion (AIC) was considered to select the best QTL model that indicated the number of meta (M) QTL. The model with the lowest AIC was considered the
best fit. When the n model was the most likely model,
the meta-analysis was performed again, choosing a subset of the QTL. The MQTL were obtained from the midpoint positions of the overlapping QTL.
3. Results
3.1. Evaluation of the Phenotypic Data. The analysis of the phenotypic data revealed that the two parents were clearly different in terms of the size of the shoot. “Molise Colli” was characterized by greater shoot height and dry weight,
compared to“Simeto.” This was expected, as “Molise Colli”
is derived from an accession of T. dicoccum, while“Simeto”
is a modern durum wheat variety. This difference in growth
between these two genotypes was also observed for the root
structure. There was a significant difference between these
parents for root dry weight (“Molise Colli,” 46.7 mg;
“Simeto,” 33.9 mg). These data indicated that “Molise Colli”
is characterized by superior growth with respect to“Simeto”
for both the aerial and root parts, even if the differences
observed for the root lengths, surface areas, diameters, volumes, and numbers of tips were not significantly different. For the segregating population, there was a large range of variation for all of the examined traits, with sig-nificant differences across the RILs (Table 2; Supplementary Tables S2 and S3).
In the PCA, the first two principal components (PCs)
explained about 65% of the total variation among the 138 genotypes evaluated for 50 morphological root traits (Figure 1). According to the factor loadings, PC1 was posi-tively correlated with root volume, length, and surface area
Table 1: Details of all of the traits for which a QTL was identified in the present study.
Trait Unit Abbreviation
Crossing number — C
Shoot number per plant — Shoots
Plant height cm PH
Shoot dry weight g SDW
Root dry weight g RDW
Length cm L
Length, diameter class 0.0 < d ≤ 0.5 mm cm L1
Length, diameter class 0.5 < d ≤ 1.0 mm cm L2
Length, diameter class 1.0 < d ≤ 1.5 mm cm L3
Length, diameter class 1.5 < d ≤ 2.0 mm cm L4
Length, diameter class 2.0 < d ≤ 2.5 mm cm L5
Length, diameter class 2.5 < d ≤ 3.0 mm cm L6
Length, diameter class 4.0 < d ≤ 4.5 mm cm L9
Surface area cm2 SA
Surface area, diameter class 0.0 < d ≤ 0.5 mm cm2 SA1
Surface area, diameter class 0.5 < d ≤ 1.0 mm cm2 SA2
Surface area, diameter class 1.0 < d ≤ 1.5 mm cm2 SA3
Surface area, diameter class 1.5 < d ≤ 2.0 mm cm2 SA4
Surface area, diameter class 2.0 < d ≤ 2.5 mm cm2 SA5
Surface area, diameter class 2.5 < d ≤ 3.0 mm cm2 SA6
Surface area, diameter class 4.0 < d ≤ 4.5 mm cm2 SA9
Volume cm3 V
Volume, diameter class 0.0 < d ≤ 0.5 mm cm3 V1
Volume, diameter class 0.5 < d ≤ 1.0 mm cm3 V2
Volume, diameter class 1.0 < d ≤ 1.5 mm cm3 V3
Volume, diameter class 1.5 < d ≤ 2.0 mm cm3 V4
Volume, diameter class 2.0 < d ≤ 2.5 mm cm3 V5
Volume, diameter class 2.5 < d ≤ 3.0 mm cm3 V6
Volume, diameter class 4.0 < d ≤ 4.5 mm cm3 V9
Number of tips — T
Number of tips, diameter class 0.0 < d ≤ 0.5 mm — T1
Number of tips, diameter class 0.5 < d ≤ 1.0 mm — T2
Number of tips, diameter class 1.5 < d ≤ 2.0 mm — T4
Number of tips, diameter class 2.0 < d ≤ 2.5 mm — T5
Number of tips, diameter class 2.5 < d ≤ 3.0 mm — T6
in the root-diameter classes from 0.5 mm to 3.0 mm. PC2 was negatively associated with the number of crossings and the root length in the root-diameter classes from 0.0 mm to
0.5 mm. In particular, “Molise Colli” showed higher values
for all root traits in the root-diameter classes 0.0 mm to 2.0 mm, for which the most significant differences were
Table 2: Phenotypic variations among the parental lines and RILs from the “Simeto” × “Molise Colli” population for the shoot and root traits.
Trait Units “Molise Colli” “Simeto” Mean Min. Max. Range ±SD CV (%) H LSD0.05
DAS Days 23.30 22.80 24.70 19.80 34.00 14.30 2.70 11.00 0.35 4.22 PH cm 60.90 39.40 53.40 36.20 71.20 35.00 8.10 15.20 0.76 6.09 Shoots N 1.75 1.00 1.58 1.00 2.25 1.25 0.38 24.35 0.29 0.66 SDW mg 212.90 162.60 221.70 121.00 380.90 260.00 56.50 25.50 0.63 0.06 RDW mg 46.70 33.90 51.50 13.50 106.60 93.10 18.30 35.50 0.27 0.03 L cm 1317.20 1096.20 1355.00 377.20 2439.40 2062.10 395.40 29.20 0.35 616.70 SA cm2 135.40 108.10 128.20 33.90 234.00 200.20 40.30 31.40 0.47 52.30 D mm 0.33 0.31 0.30 0.23 0.38 0.15 0.03 9.10 0.08 0.07 F N 4546.25 3270.00 5352.22 994.00 12172.75 11178.75 2352.46 43.95 0.34 3747.18 V cm3 1.12 0.86 0.99 0.25 2.22 1.96 0.36 36.50 0.45 0.48 T N 1860.30 1513.80 1990.10 679.00 3303.80 2624.80 597.50 30.00 0.35 929.20
Min.: minimum value; max.: maximum value; SD: standard deviation; CV: coefficient of variation; H: broad sense heritability; LSD: least significant difference; DAS: days after sowing; PH: plant height; shoots: number of shoots per plant; SDW: shoot dry weight; RDW: root dry weight; L: length; SA: surface area; D: diameter; F: number of forks; V: volume; T: number of tips.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 19 20 22 23 26 27 29 30 31 32 35 37 38 40 43 48 49 50 52 54 55 56 57 59 60 61 63 67 68 70 72 74 76 78 79 80 81 83 88 89 90 91 94 95 96 99 137B 172B 188B 105 106 107 113 114 115 117 121 122 124 125 126 127 128 130 131 133 137B 138 139 142 144 145 148 149 150 151 152 153 154 156 158 159 160 161 163 164 165 166 167 170 172B 174 177 178 181 182 185 187 188B 189 190 191 192 193 194 195 196 197 198 199 201 203 204 205 206 207 208 209 210 211 92A Molise Simeto −10 −5 0 5 10 15 20 25 PC 1: 48.45% −10 −5 0 5 10 PC 2: 16.08%
Figure 1: Principal component analysis score plot of the first two principal components of the parental lines and RILs from the durum wheat “Simeto” × “Molise Colli” population for the root traits.
observed in terms of total variability, while “Simeto” was phenotypically superior for root traits in the larger root-diameter classes (Supplementary Table S2). These traits can be considered as key characteristics for the estimation of the genetic diversity in a durum wheat population. Due to the high phenotypic variation, the scatter diagram showed wide dispersion along both of the PC axes and evident and
significant groups of genotypes with different root
architec-tures (Figure 1).
Moreover, correlation analysis was carried out consider-ing the root traits evaluated in the present study and the traits related to seed size and morphology that were previously analyzed [57]. In particular, the means over 2 years of field evaluation were considered, and the most significant correlations were between 1000-kernel weight and seed area on the one hand and a number of root traits on the other (Supplementary Table S4).
3.2. QTL Mapping for Root Traits in the Durum Wheat “Simeto” × “Molise Colli” RIL Population. The linkage map used for the QTL analysis included SSR and SNP markers for a total of 9040 markers that covered 2879.3 cM [57]. A total of 61 QTL covering 17 chromosomal regions were identified in the present study, which were located on chromosomes 1B, 2A, 3A, 4B, 5B, 6A, 6B, and 7B (Table 3). Some of these QTL controlled the above-ground biomass (e.g., number of shoots, plant height, and shoot dry weight),
and for all of these, the “Molise Colli” allele effect was
positive, as indicated by the negative sign of the additive effects. The analysis with the markers MR1 and
BF-MR2 allowed “Simeto” to be assigned by the Rht-B1b
allele, while “Molise Colli” was characterized by the
Rht-B1a allele. The scoring of the markers across the segregat-ing population led to locate this locus on the SMC genetic map (Figure 2). The region on chromosome 4B that corre-sponds to the Rht-B1 region was of particular interest; this controlled plant height, shoot dry weight, and a number of root morphological traits, most of which were in the smallest root-diameter classes. The LODs were very high for the shoot
traits (12.1–21.6), with the explained variability between 52%
for plant height and 34% for shoot dry weight. For the root traits controlled by this QTL, the LODs were between 3.1 for root volume class 3 and 7.4 for root surface area. The observed variability explained by this QTL was from
10% to 22%. The highest R2 values were observed for root
surface area and volume in root-diameter class 1. All of the root traits explained by this QTL were in root-diameter classes 1, 2, and 3, except for root volume and surface area, which were in class 6. As well as this region on chromosome 4B, other QTL involved in the control of shoot traits were identified: QTL6 for the number of shoots per plant and number of root tips in root-diameter class 2 was located on chromosome 2A and explained 17% of the observed variability, with a LOD of 5.4. The allele of “Molise Colli” was effective in increasing the trait. The
same allelic effect was found for QTL7 on chromosome
3A and QTL13 on chromosome 6A, which controlled plant height (QTL7) and plant height and leaf dry weight (QTL13). The LODs were between 3.4 and 5.2 for these
QTL, which explained from 11% to 17% of the observed phenotypic variability.
All of the other QTL identified in the present study were specifically involved in the control of root traits. In some cases, these QTL controlled only a single trait, as for chromo-some region 5 on chromochromo-some 2A for root volume and chro-mosome region 2 on chrochro-mosome 1B for the number of root
tips in root-diameter class 2. For the allelic effects, the effect
of the “Molise Colli” allele was positive for QTL2, and the
effect of the “Simeto” allele was positive for a QTL identified in chromosome region 5. Most of the QTL identified in the present study showed effects on many different root traits, but only for specific root-diameter classes. The QTL identi-fied in chromosome regions 11, 14, and 15 on chromosomes 4B, 6A, and 6B, respectively, were involved in the control of various root traits and, in particular, for root-diameter class 1. The QTL mapped to chromosome region 10 were also on chromosome 4B, and as well as the number of forks, they controlled the root lengths, volumes, and surface area as in
root-diameter class 2. A positive effect of the “Molise Colli”
allele was observed for all of these QTL. QTL in chromosome region 16 on chromosome 6B explained 9% to 12% of the observed phenotypic variability for root length, volume, and surface area for root-diameter classes 4 to 6. In this case, the values of these traits were increased by the allele of “Simeto.” The same allelic effect was observed for QTL in region 17 on chromosome 7B, which explained 11% of the observed variability for length, surface area, and volume, but only for root-diameter class 9.
3.3. Meta-QTL Analysis. A number of studies have reported QTL for root traits in wheat based on reliable information,
such as R2, confidence intervals, and common markers (see
Supplementary Table S1). To compare the QTL regions in the SMC genetic map with those reported in the literature and to identify the precise consensus QTL, MQTL analysis was carried out. A very dense consensus map comprising the A and B genomes was used for this meta-analysis. This was composed of >40,000 markers and spanned a total map
length of 2791 cM. More details about the final consensus
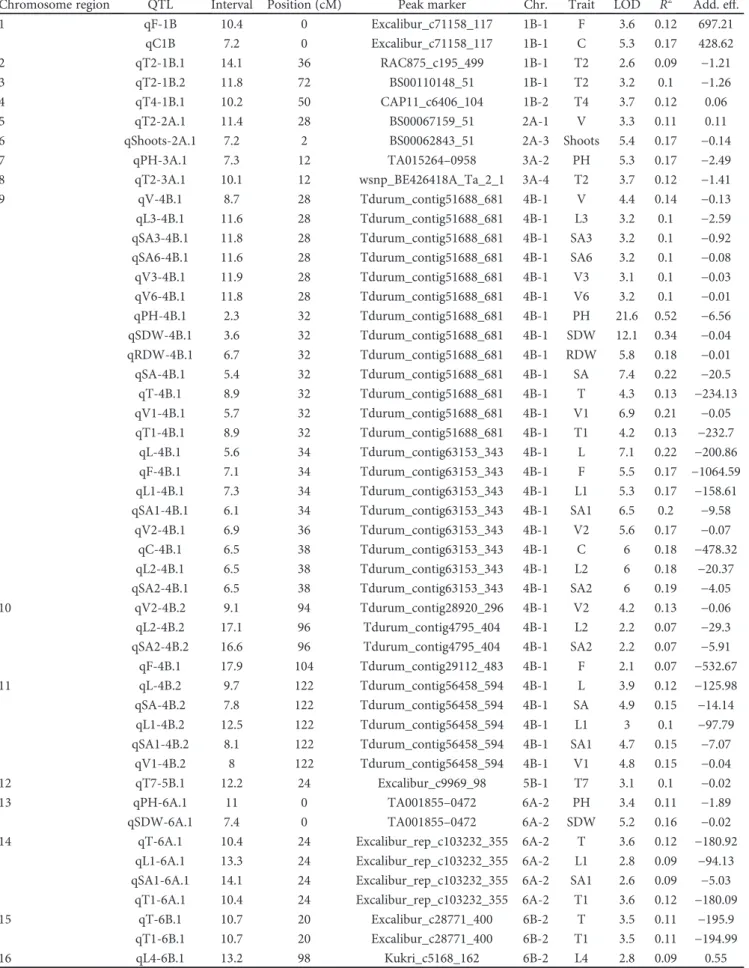
map will be the aim of a future study, such as the number of markers per chromosome and the mean marker distances. Here, only the chromosomes involved with the QTL identified in the SMC genetic map are reported, along with the data on the MQTL (Figure 2).
The meta-analysis was launched on 100 QTL for a num-ber of traits linked to the growth and morphology of the root structure retrieved from the literature and the 17 QTL regions identified in the present study, which resulted in 34 MQTL. The MQTL merged from two to eight individual QTL. The MQTL are reported in Table 4, along with the AICs, confidence intervals, flanking markers, and number of initial QTL involved. Twenty-nine out of the initial 100 QTL remained as singletons, their numbers per chromosome ranged from 1 (3A, 5B, 7B) to 2 (1B), and some of these were found only in the SMC (Figure 2, Table 4). The 95%
confidence intervals of the MQTL varied from 0.5 cM to
30.5 cM, with a mean of 5.6 cM. The MQTL were on chromo-somes 1B, 2A, 3A, 4B, 5B, 6A, 6B, and 7B. All of the MQTL
Table 3: QTL detected in the “Simeto” × “Molise Colli” RIL populations for the evaluated traits.
Chromosome region QTL Interval Position (cM) Peak marker Chr. Trait LOD R2 Add. eff.
1 qF-1B 10.4 0 Excalibur_c71158_117 1B-1 F 3.6 0.12 697.21 qC1B 7.2 0 Excalibur_c71158_117 1B-1 C 5.3 0.17 428.62 2 qT2-1B.1 14.1 36 RAC875_c195_499 1B-1 T2 2.6 0.09 −1.21 3 qT2-1B.2 11.8 72 BS00110148_51 1B-1 T2 3.2 0.1 −1.26 4 qT4-1B.1 10.2 50 CAP11_c6406_104 1B-2 T4 3.7 0.12 0.06 5 qT2-2A.1 11.4 28 BS00067159_51 2A-1 V 3.3 0.11 0.11
6 qShoots-2A.1 7.2 2 BS00062843_51 2A-3 Shoots 5.4 0.17 −0.14
7 qPH-3A.1 7.3 12 TA015264–0958 3A-2 PH 5.3 0.17 −2.49
8 qT2-3A.1 10.1 12 wsnp_BE426418A_Ta_2_1 3A-4 T2 3.7 0.12 −1.41
9 qV-4B.1 8.7 28 Tdurum_contig51688_681 4B-1 V 4.4 0.14 −0.13
qL3-4B.1 11.6 28 Tdurum_contig51688_681 4B-1 L3 3.2 0.1 −2.59
qSA3-4B.1 11.8 28 Tdurum_contig51688_681 4B-1 SA3 3.2 0.1 −0.92
qSA6-4B.1 11.6 28 Tdurum_contig51688_681 4B-1 SA6 3.2 0.1 −0.08
qV3-4B.1 11.9 28 Tdurum_contig51688_681 4B-1 V3 3.1 0.1 −0.03 qV6-4B.1 11.8 28 Tdurum_contig51688_681 4B-1 V6 3.2 0.1 −0.01 qPH-4B.1 2.3 32 Tdurum_contig51688_681 4B-1 PH 21.6 0.52 −6.56 qSDW-4B.1 3.6 32 Tdurum_contig51688_681 4B-1 SDW 12.1 0.34 −0.04 qRDW-4B.1 6.7 32 Tdurum_contig51688_681 4B-1 RDW 5.8 0.18 −0.01 qSA-4B.1 5.4 32 Tdurum_contig51688_681 4B-1 SA 7.4 0.22 −20.5 qT-4B.1 8.9 32 Tdurum_contig51688_681 4B-1 T 4.3 0.13 −234.13 qV1-4B.1 5.7 32 Tdurum_contig51688_681 4B-1 V1 6.9 0.21 −0.05 qT1-4B.1 8.9 32 Tdurum_contig51688_681 4B-1 T1 4.2 0.13 −232.7 qL-4B.1 5.6 34 Tdurum_contig63153_343 4B-1 L 7.1 0.22 −200.86 qF-4B.1 7.1 34 Tdurum_contig63153_343 4B-1 F 5.5 0.17 −1064.59 qL1-4B.1 7.3 34 Tdurum_contig63153_343 4B-1 L1 5.3 0.17 −158.61
qSA1-4B.1 6.1 34 Tdurum_contig63153_343 4B-1 SA1 6.5 0.2 −9.58
qV2-4B.1 6.9 36 Tdurum_contig63153_343 4B-1 V2 5.6 0.17 −0.07
qC-4B.1 6.5 38 Tdurum_contig63153_343 4B-1 C 6 0.18 −478.32
qL2-4B.1 6.5 38 Tdurum_contig63153_343 4B-1 L2 6 0.18 −20.37
qSA2-4B.1 6.5 38 Tdurum_contig63153_343 4B-1 SA2 6 0.19 −4.05
10 qV2-4B.2 9.1 94 Tdurum_contig28920_296 4B-1 V2 4.2 0.13 −0.06
qL2-4B.2 17.1 96 Tdurum_contig4795_404 4B-1 L2 2.2 0.07 −29.3
qSA2-4B.2 16.6 96 Tdurum_contig4795_404 4B-1 SA2 2.2 0.07 −5.91
qF-4B.1 17.9 104 Tdurum_contig29112_483 4B-1 F 2.1 0.07 −532.67
11 qL-4B.2 9.7 122 Tdurum_contig56458_594 4B-1 L 3.9 0.12 −125.98
qSA-4B.2 7.8 122 Tdurum_contig56458_594 4B-1 SA 4.9 0.15 −14.14
qL1-4B.2 12.5 122 Tdurum_contig56458_594 4B-1 L1 3 0.1 −97.79
qSA1-4B.2 8.1 122 Tdurum_contig56458_594 4B-1 SA1 4.7 0.15 −7.07
qV1-4B.2 8 122 Tdurum_contig56458_594 4B-1 V1 4.8 0.15 −0.04
12 qT7-5B.1 12.2 24 Excalibur_c9969_98 5B-1 T7 3.1 0.1 −0.02
13 qPH-6A.1 11 0 TA001855–0472 6A-2 PH 3.4 0.11 −1.89
qSDW-6A.1 7.4 0 TA001855–0472 6A-2 SDW 5.2 0.16 −0.02
14 qT-6A.1 10.4 24 Excalibur_rep_c103232_355 6A-2 T 3.6 0.12 −180.92
qL1-6A.1 13.3 24 Excalibur_rep_c103232_355 6A-2 L1 2.8 0.09 −94.13
qSA1-6A.1 14.1 24 Excalibur_rep_c103232_355 6A-2 SA1 2.6 0.09 −5.03
qT1-6A.1 10.4 24 Excalibur_rep_c103232_355 6A-2 T1 3.6 0.12 −180.09
15 qT-6B.1 10.7 20 Excalibur_c28771_400 6B-2 T 3.5 0.11 −195.9
qT1-6B.1 10.7 20 Excalibur_c28771_400 6B-2 T1 3.5 0.11 −194.99
identified corresponded to chromosomal regions that are involved in the control of a number of root traits. In some cases, these resulted from individual QTL, each of which is involved in the control of a different trait. As an example, MQTL22 was mapped to chromosome 6A and merged three individual QTL for root numbers, lateral roots, and root dry weights. In the other cases, the MQTL merged two or more QTL that explained the same trait, for example, MQTL23 on chromosome 6A merged two individual QTL for root dry weight and one QTL for total root length, and one more that explained root length, surface area, volume, and tip number. Based on the molecular markers mapped to both chromosomes 6A and 6B, a homoeologous relationship can be established between MQTL26 (6A) and 30 (6B) and between MQTL27 (6A) and 31 (6B). In particular, MQTL27 and MQTL31 are both involved in the control of root length.
Based on this MQTL analysis, three QTL identified in the
SMC in the present study did not correspond to previously published QTL. Two QTL were mapped to chromosomes 3A and 5B and were related to the number of root tips,
although in different root-diameter classes. The third QTL,
on chromosome 7B, was involved in the control of root length, surface area, and volume, although only in the largest root-diameter class.
4. Discussion
A number of methods have been described for phenotypic evaluation of root morphology under controlled conditions for numerous samples, as required for genetic analyses (e.g., [2–5, 35]). In the present study, a method in which the plants were grown in soil mixed with sand was used, to have more reliable data. The use of this growth substrate and the growth stage considered allowed us to carry out an evaluation of the root system that is independent of the effect of seed weight.
For bread and durum wheat, linkage and association mapping studies have both been carried out to identify chro-mosome regions involved in the control of root traits. In most studies, elite and old cultivars were used, and although good phenotypic variability has been observed [31, 35, 37], it can be useful to consider more genetically diverse genotypes
to search for novel haplotypes that control root architecture. On this basis, a biparental population derived from a durum wheat elite cultivar and a T. dicoccum accession represents a valuable resource to identify novel loci of interest for root
traits. The parents of this SMC,“Simeto” and “Molise Colli,”
are very different in terms of plant morphology, for both the aerial and below-ground organs, and this can provide indica-tions of the relaindica-tionships between plant height and the devel-opment of the root system. Moreover, the two parents are
also different in terms of seed size and morphology, and in
a previous study, a genetic map with more than 9000 SNP markers was used to identify QTL for seed morphology [57]. When evaluated at Zadoks stage 15, the two parents were clearly different for plant height, shoot dry weight,
and root dry weight, whereby“Molise Colli” showed greater
growth with respect to“Simeto” for the whole plant. When
specific root traits were considered, in all cases, “Molise
Colli” had higher phenotypic expression compared to
“Simeto,” although this difference did not reach significance.
Significant differences were observed across the RIL
popula-tion, with large and transgressive variations for all of the traits examined. This indicates that both genotypes have loci that contribute to the development of the root structure.
The MQTL analysis was carried out to compare the genetic positions of the QTL identified in the present study with those of previously published QTL in wheat. An inte-grated map that contained different types of molecular markers was used to project the known QTL, which is of par-ticular importance as the SMC genetic map which nearly
contains only SNPs from the Infinium 90K wheat assay.
Some of the QTL identified in the present study were seen
for chromosome regions in which a MQTL was present, as for MQTL1 (chromosome 1B), MQTL2 (chromosome 2A), MQTL3 (chromosome 3A), MQTL9 (chromosome 6A), MQTL11 and MQTL12 (chromosome 6B), and MQTL13 (chromosome 7B). For MQTL2 and MQTL3, the QTL for the number of shoots per plant and for plant height, respec-tively, were coincident with the QTL previously reported for root traits. Closely linked genes or a single locus with
pleio-tropic effects might be responsible for these different traits.
Considering the results of this MQTL analysis and the studies
Table 3: Continued.
Chromosome region QTL Interval Position (cM) Peak marker Chr. Trait LOD R2 Add. eff.
qSA4-6B.1 13 98 Kukri_c5168_162 6B-2 SA4 2.8 0.09 0.29
qSA6-6B.1 13.3 98 Kukri_c5168_162 6B-2 SA6 2.8 0.09 0.07
qV4-6B.1 13 98 Kukri_c5168_162 6B-2 V4 2.9 0.09 0.01
qV6-6B.1 13.5 98 Kukri_c5168_162 6B-2 V6 2.8 0.09 0.01
qL5-6B.1 10.2 100 CAP7_c3697_87 6B-2 L5 3.7 0.12 0.23
qL6-6B.1 13.5 100 CAP7_c3697_87 6B-2 L6 2.8 0.09 0.08
qSA5-6B.1 10.2 100 CAP7_c3697_87 6B-2 SA5 3.7 0.12 0.15
qV5-6B.1 10.1 100 CAP7_c3697_87 6B-2 V5 3.7 0.12 0.01
17 qL9-7B.1 11.3 14 RAC875_c23521_589 7B-1 L9 3.3 0.11 0.01
qSA9-7B.1 11.3 14 RAC875_c23521_589 7B-1 SA9 3.3 0.11 0.01
qV9-7B.1 11.1 14 RAC875_c23521_589 7B-1 V9 3.4 0.11 0.001
wPt-8328 0.0 Xwmc382 1.3 Xwmc326c 4.0 Excalibur_c59428_237 5.2 Xbarc212 5.6 BS00105593_51 6.4 CAP7_c4676_94 7.8 BS00080570_51 9.8 D_contig79877_194 10.6 wPt-2147 11.3 Tdurum_contig49144_321 12.8 D_contig71899_62 13.2 Jagger_c5341_126 15.7 Xwmc326b 16.0 BS00014897_51 16.7 Tdurum_contig30824_197 16.9 Tdurum_contig55029_963 17.4 RAC875_c1968_103 18.3 wPt-8925 19.1 Excalibur_c12980_2392 20.0 RAC875_c8069_1874 20.5 RAC875_c57889_55 21.6 Xmag3807 22.5 wPt-4197 23.0 Ex_c12425_506 25.4 BobWhite_c10673_330 26.4 wPt-4984 27.4 wPt-9672 28.3 RAC875_c2300_1021 29.3 Xgwm512 30.2 BS00065192_51 30.8 Xwmc728 32.0 Tdurum_contig74742_224 34.0 BS00107263_51 35.2 Xwmc630b 37.5 Excalibur_rep_c106338_424 39.7 Xwmc602 41.1 wPt-1368 42.1 Excalibur_c21663_145 43.9 BS00067159_51 44.0 Xwmc149 45.7 wPt-5839 46.6 BS00067684_51 47.6 BS00069752_51 48.5 RAC875_rep_c113106_56 50.6 Kukri_c63732_593 52.4 wPt-3896 53.7 CAP11_c2293_200 54.0 BobWhite_rep_c51499_280 61.7 BS00068139_51 62.9 BS00084333_51 63.2 BS00075855_51 64.4 BobWhite_c19433_329 65.4 BS00070797_51 66.0 RAC875_rep_c78744_228 67.1 BS00076693_51 67.9 BS00065276_51 68.3 Excalibur_c43811_527 69.3 Excalibur_rep_c102244_1103 70.9 D_GA8KES401BVP4P_43 73.8 BobWhite_c26374_339 74.3 Xwmc826 75.0 BobWhite_c30988_361 76.0 Tdurum_contig32692_271 76.6 wsnp_Ex_c5412_9564346 77.3 Tdurum_contig44316_486 77.9 Ra_c58279_684 78.4 Excalibur_c54892_188 79.0 wsnp_Ex_c30481_39394826 79.7 wPt-7285 80.2 Xgwm296 81.3 BS00044274_51 82.3 wPt-4201 83.2 Tdurum_contig62138_556 84.1 Xwmc453 84.8 Xwmc51d 85.6 wPt-9320 86.1 Excalibur_c25235_1289 88.0 wPt-3037 88.5 wPt-2435 89.3 Xwmc522 90.1 Xwmc32 91.4 RAC875_c17787_274 92.0 BS00070693_51 92.7 Excalibur_rep_c103108_709 93.9 Xwmc474 94.1 TA004602-1630 94.8 Xwmc792 95.4 Tdurum_contig93115_517 96.1 BS00078612_51 97.9 Excalibur_c37649_125 98.8 Xgwm275 99.4 BS00055933_51 100.0 Xbcd855 100.5 Xbcd1709 101.8 Xgwm558 102.1 D_contig15270_478 102.8 Tdurum_contig10579_413 103.5 Xgwm95 104.2 GENE-1350_36 104.5 Xwmc632 105.4 Xwmc455 106.2 BS00062843_51 106.6 Xgwm372 107.1 Xbarc353 107.9 Xwmc819 108.2 BS00078116_51 108.8 BS00022301_51 109.0 Xcfa2263 109.6 Xbarc5 110.1 RFL_Contig4517_961 111.5 IAAV4464 112.5 BS00070004_51 113.7 Xgwm445 114.6 BS00000297_51 115.8 Excalibur_c40617_983 116.4 Xgwm817 117.1 BS00029332_51 117.6 wPt-5865 118.8 BobWhite_c2058_367 120.3 BAC875_c77816_365 121.6 BS00107804_51 122.3 Excalibur_c7282_285 122.9 Excalibur_c3171_416 123.4 BobWhite_c17403_635 124.0 BS00065865_51 124.9 RAC875_c9691_870 125.8 Tdurum_contig11678_289 126.8 BS00080751_51 127.4 Ku_c25175_1248 127.8 RAC875_c16333_340 128.0 Xwmc109 129.0 BS00079611_51 129.3 RAC875_rep_c107961_348 130.8 GENE-1908_331 131.8 wsnp_Ex_c14953_23104041 132.4 BS00024921_51 132.8 BS00067907_51 133.1 Kukri_c43875_671 133.8 BS00023048_51 134.0 RAC875_c104160_61 135.1 BS00065366_51 135.7 BS00001260_51 136.0 Excalibur_c33040_447 139.0 wPt-3244 139.6 Tdurum_contig93508_119 139.9 BS00077768_51 140.2 RAC875_c25848_122 140.7 wPt-7056 141.2 Excalibur_c44487_642 141.8 RFL_Contig785_1156 142.0 Excalibur_c8009_325 142.7 BS00022409_51 143.0 BobWhite_c17783_174 143.6 BS00110516_51 144.0 RAC875_c9523_328 144.5 RAC875_c1758_373 146.8 RAC875_c4609_1756 148.4 BS00108288_51 149.2 TA004785-1734 149.9 BS00096927_51 150.0 BS00102709_51 150.5 Excalibur_c7971_985 150.7 Excalibur_c21117_300 151.4 Tdurum_contig92425_1612 152.0 Excalibur_c63622_125 152.7 Xgwm356 153.1 Xgwm761 153.5 Xgwm526 153.9 RAC875_c1789_253 154.0 Kukri_c41234_126 155.0 BS00022252_51 155.4 Xwmc658 156.9 Xwmc198 157.3 Xcfd86 157.7 Xwmc181 159.4 Excalibur_c15671_87 160.1 wPt-669355 160.7 tPt-3136 161.5 Tdurum_contig92425_1324 162.0 wPt-9586 163.6 Excalibur_c21501_237 164.6 wPt-5850 165.1 Excalibur_c24213_314 165.5 Xgwm1067 165.8 wPt-1615 166.2 RFL_Contig5277_1283 166.7 BS00091763_51 167.0 Tdurum_contig92425_431 168.0 GENE-0762_808 168.3 Xbarc76 171.9 Ku_c105361_414 173.1 wPt-3728 174.9 Xgwm382 175.3 Excalibur_c12916_123 176.9 BS00095525_51 177.3 IAAV5706 178.0 Xgwm311 179.2 Tdurum_contig42013_538 179.5 BS00022510_51 180.1 RFL_Contig2972_2819 180.5 Tdurum_contig50720_204 182.2 Excalibur_c27531_86 184.2 Ku_c68139_836 185.8 2A D_GDEEGVY02FU4W8_119a 0.0 CAP8_c1361_367a 11.0 Tdurum_contig84762_258a 12.1 CAP11_c7974_175a 13.4 Xbarc57 14.6 Excalibur_c11079_101a 14.9 Xwmc206c 16.0 BobWhite_rep_c49374_348 17.9 wPt-3094 18.6 Excalibur_c60530_113 20.7 RAC875_rep_c77067_347 21.1 BS00048491_51 22.7 CAP12_c2940_354 23.4 RAC875_rep_c111243_125 24.2 BS00110922_51 25.3 Excalibur_rep_c109599_89 26.0 BS00073063_51 27.6 GENE-1989_800 28.6 BS00064422_51 29.5 wsnp_Ku_c7060_12212702 31.2 Kukri_c96747_274a 32.3 RAC875_c371_251a 33.7 BobWhite_c29706_369 34.5 Excalibur_rep_c80735_81 35.5 wsnp_Ra_c9738_16174002 38.4 Tdurum_contig75764_146 40.5 BobWhite_c48246_303 42.6 wsnp_Ex_c8409_14170476 44.3 wsnp_Ex_c55051_57706127 47.2 RAC875_c17393_553 48.0 wsnp_BE497169B_Ta_2_1a 49.1 RAC875_c16583_1259 53.3 tplb0056l18_796 54.9 tplb0056l18_622 55.1 wsnp_JD_rep_c63739_40660910 55.6 BobWhite_c2448_96 56.2 Kukri_c63647_1522 59.0 wsnp_CAP12_c2692_1286812 60.2 BS00065382_51 61.1 CAP8_c1702_71 61.9 Ra_c31046_1652 63.5 Excalibur_c27972_654 63.9 Kukri_c32377_399 64.3 Excalibur_c34472_211 66.5 Tdurum_contig99640_243 67.5 Ku_c8627_745 68.7 RAC875_c10628_941 71.5 wsnp_Ex_c33765_42199371 73.2 RAC875_c14060_1935 75.1 Excalibur_c12446_155 75.6 RAC875_rep_c109701_76 76.0 Tdurum_contig45380_92 76.5 RAC875_c2292_402 77.3 wPt-7217 77.9 Tdurum_contig86206_149 78.1 RAC875_c50864_1921 79.8 wsnp_Ex_c57322_59084809 80.6 wPt-2813 81.6 Tdurum_contig14694_211 82.2 wsnp_Ku_c40218_48484410 83.9 Tdurum_contig44029_158 84.5 Tdurum_contig60631_336 85.0 Xgwm5b 85.5 RAC875_rep_c100137_586 86.0 TA002997-0276 86.2 wsnp_Ex_c3478_6369892 86.6 wPt-7608 87.4 wsnp_Ex_c14420_22402673 87.7 Tdurum_contig28100_112 88.0 Kukri_rep_c71128_1811 88.9 wsnp_Ex_rep_c66865_65262612 89.2 wsnp_JD_c5699_6859527 89.5 Xwmc527b 90.0 Excalibur_c11807_106 90.5 Tdurum_contig49981_703 90.7 Excalibur_c61765_220a 91.1 wsnp_Ex_rep_c66685_65003254 91.7 Xbarc45 92.4 wPt-2698 92.9 Excalibur_rep_c107798_68 93.0 wPt-1596 93.4 Xwmc664 93.7 wPt-6891 93.9 wPt-4077 94.2 Xwmc505a 94.6 Xgwm720 94.7 Xbarc356 95.0 Xcfd193a 95.2 Xwmc489c 96.0 Kukri_c60531_131 97.3 Kukri_rep_c111139_338 97.8 wsnp_Ex_c18223_27035083 98.4 Ra_c18366_203 98.7 Xwmc695a 99.4 Xwmc269b 100.8 wsnp_Ku_c18497_27803432 101.0 BS00074617_51 102.0 Tdurum_contig76296_461 102.7 Tdurum_contig42827_1693 103.3 Excalibur_c7544_288 103.6 wsnp_Ex_c4923_8767234 104.9 Excalibur_rep_c105978_544 105.7 BS00078669_51 106.1 BS00048355_51 106.8 Kukri_rep_c116421_403a 107.1 Xwmc264a 108.2 RAC875_c57977_121b 109.3 Excalibur_c24402_471 109.6 Tdurum_contig9912_228 109.9 Xwmc169 110.4 Ra_c1619_432 111.3 Excalibur_c39508_88 111.6 tplb0046k04_484 112.9 wsnp_Ku_c8400_14280021 113.2 BS00084158_51 113.9 Tdurum_contig8584_354 114.6 BS00041339_51 115.6 F129 115.9 Tdurum_contig49518_1002 116.3 Xgwm494b 117.0 wPt-5943a 117.6 Kukri_c49798_300 119.5 wsnp_Ex_c1894_3575749 120.2 RAC875_c15003_377 122.7 wsnp_Ex_c12341_19693090 123.1 IAAV5984 124.3 KukBi_c10751_264 125.1 Tdurum_contig8365_319 126.7 Kukri_c15151_436 127.8 Xgwm751a 128.4 RAC875_rep_c90531_283 130.2 RAC875_c15109_106 131.0 Xwmc96a 131.4 Xbarc69 133.6 Ku_c1255_1107 136.2 Excalibur_c3429_652b 138.1 Hu_c1255_862 138.9 wPt-4407 140.2 Xwmc153 141.3 wsnp_Ku_c10468_17301042a 141.9 Ku_c6126_1140 142.6 Xwmc173a 143.3 Excalibur_c640_464b 143.8 Xwmc559 144.0 GENE-1918_152 144.6 Excalibur_c3969_1208 145.6 Xwmc215a 146.1 RAC875_c52195_324 146.9 Kukri_c29993_593 147.9 Xgwm155 148.2 KukBi_c6645_570 149.3 Xcfa2076 150.2 BobWhite_c31562_95 151.8 Xgwm156c 152.1 GENE-3665_61 153.6 Tdurum_contig55841_351a 154.2 RAC875_c18230_901 154.7 Xbarc51 155.3 Xbarc19 156.6 wPt-1864 157.2 Kukri_c2596_146 158.4 wsnp_BE604885A_Ta_2_1 159.7 TduBum_contiA10544_92 160.1 Tdurum_contig28518_122 161.5 Excalibur_rep_c76177_243 162.1 Xgwm480 164.1 wPt-2659 165.2 wPt-3816 166.5 tPt-7492 167.3 BS00108976_51 168.1 RFL_Contig114_878 168.9 Xwmc206d 172.1 BS00059618_51a 173.2 RAC875_c3084_415 174.1 Excalibur_c77321_69 175.3 wPt-9422 176.5 wPt-9238 176.7 Excalibur_c361_1321 176.9 Excalibur_c77321_196a 177.4 wPt-2492 178.0 Xbarc314 178.6 Tdurum_contig38751_317 179.5 wPt-3978 179.8 Tdurum_contig50577_620 180.3 TC77302 180.4 Excalibur_c90478_62 180.6 Excalibur_c71730_105a 181.1 wsnp_Ex_rep_c66274_64426834 182.1 Tdurum_contig13646_292 182.7 wPt-2144 182.9 wsnp_Ex_c11229_18163892 184.8 wsnp_Ex_c28679_37784735 185.4 wsnp_Ra_c66411_64796843 187.0 Xgwm162a 187.4 Kukri_rep_c70441_132 188.4 Excalibur_rep_c114569_56 189.5 RAC875_c46236_214 190.3 Tdurum_contig75336_518 191.4 Tdurum_contig51914_740 193.8 RAC875_c20905_383 194.3 Tdurum_contig16865_174 195.1 Tdurum_contig10022_413 197.2 GENE-1910_570 207.3 3A BS00071333_51 0.0 RFL_Contig293_498 0.7 Excalibur_c10065_411 2.1 RAC875_c18591_175a 4.1 Excalibur_c71158_117 5.0 Kukri_c18951_853b 6.0 Kukri_c18951_493 6.6 RAC875_c24163_155 9.1 Tdurum_contig28703_293 10.1 Excalibur_c51033_173 11.0 Kukri_c5384_86 11.5 BS00023084_51a 14.9 Excalibur_rep_c107678_98 16.7 Tdurum_contig403_122 17.7 GENE-1322_33 18.3 D_GB5Y7FA01C2ERV_339 19.4 BS00015608_51b 20.5 wPt-0308 21.7 D_GDRF1KQ01EEUZ7_267 22.2 Excalibur_c28255_482 22.9 Excalibur_c23212_125 23.4 ExcalibuB_c71158_117 24.7 Ku_c36151_691 25.2 BS00038243_51b 26.5 wPt-304810 27.2 Kukri_c24615_823 28.7 BobWhite_c17559_105 29.5 BS00022101_51 30.4 BS00022429_51 31.0 BS00064829_51 31.6 BobWhiBe_c28295_256 32.4 BS00062122_51 32.8 wPt-1912 37.5 wsnp_Ku_c7822_13408189 39.2 wsnp_Ex_rep_c67665_66326492b 40.2 TduBum_contiA50555_944 40.7 Tdurum_contig50667_409 41.1 BobWhite_c28295_256 41.6 wPt-4811 41.8 wPt-4434 42.0 Xbarc128a 42.6 wPt-3177 43.0 Xgwm24 43.2 wPt-7529 43.7 wPt-5164 44.1 Xbarc119b 44.5 Tdurum_contig83093_443 44.8 BS00066682_51 45.0 Xgwm413 45.2 wPt-7242 46.0 BobWhite_c3335_1438a 46.3 wPt-6777 46.7 wPt-3889 47.1 wsnp_Ex_c14273_22230844 47.9 Tdurum_contig44861_581 48.3 Xgwm133a 50.5 wPt-1399 51.3 BS00067043_51 51.8 Excalibur_c6726_357 53.2 Tdurum_contig9811_127 53.8 Tdurum_contig61410_467 54.1 Tdurum_contig42446_1999 56.1 Tdurum_contig60037_528 56.7 RAC875_c16247_107a 57.1 Tdurum_contig11004_688 57.8 TA005995-0488 58.0 Tdurum_contig17609_117 58.5 BobWhite_c33976_317 58.7 P4133-170 59.3 Kukri_c5861_360 60.2 RAC875_c195_499 60.7 Tdurum_contig24287_300 61.1 BS00067586_51 61.3 TA001559-0515 62.0 RAC875_c14049_376 62.3 RAC875_c25692_343 63.1 Excalibur_c56657_282 63.5 BS00065117_51 63.7 Excalibur_rep_c108957_173 64.0 RAC875_c4377_524 64.4 RAC875_c1914_942 64.6 RAC875_rep_c70022_1648 65.0 Excalibur_c2181_927 65.4 BobWhite_c4126_442 66.0 BS00031023_51 66.5 wsnp_Hu_c30982_40765254 67.3 Xgwm498 67.6 wsnp_Ku_c12464_20125626a 68.0 Xcfd59b 68.9 BS00012051_51 69.5 BS00022133_51 70.0 RAC875_c46646_549 70.8 wsnp_Ex_c31567_40338517 71.3 Xbarc120 71.5 RAC875_c8878_232 72.0 Tdurum_contig98080_171 72.7 Xbarc240b 74.0 BS00022343_51 74.3 wsnp_Ex_c1600_3051075 74.7 Tdurum_contig29363_461 75.0 BS00085546_51 75.4 BS00022742_51 76.0 IACX9510 76.5 Excalibur_c48379_116 76.8 RAC875_rep_c111001_187 77.2 Ra_c62804_1641 77.6 RAC875_c22886_261 78.1 RFL_Contig4576_702 78.9 wsnp_Ex_rep_c69766_68723140 79.7 Xbarc181 80.2 wsnp_Ex_rep_c66389_64588992 80.5 CAP8_c4697_108a 81.5 Tdurum_contig57101_505 82.4 wPt-3579 82.6 BS00081396_51 83.0 Tdurum_contig47550_699 84.2 Tdurum_contig30517_578 85.1 wsnp_Ex_c9577_15857465 86.1 Tdurum_contig13117_1316 86.8 RAC875_rep_c106876_558 87.4 wsnp_Ex_c9063_15093396a 87.8 CA651264 89.3 Ku_c1932_1583 90.0 wsnp_Ex_rep_c67299_65845319 90.3 Ku_c17871_1360 90.7 Ku_c9909_1766 91.3 BS00072289_51 93.9 Excalibur_c94658_59 95.1 BS00012029_51 95.7 Tdurum_contig85180_99 96.0 wsnp_Ku_c56140_59771244 97.1 Xbarc81 97.5 RFL_Contig2826_548 98.3 RFL_Contig5937_1677 99.8 RAC875_c291_2201 100.1 wsnp_JD_c1544_2179305 100.9 Tdurum_contig67656_58 101.5 Tdurum_contig67368_157 103.1 wPt-2257 103.4 Excalibur_c33470_175 104.3 Tdurum_contig65853_534 105.1 BS00022249_51 105.4 BS00072791_51 106.5 KukBi_c32448_268 106.9 Tdurum_contig11077_122 107.8 BS00011596_51 108.0 Excalibur_c65152_572 109.1 ExcalibuB_c35755_110 109.9 Ra_c7324_527 110.4 BobWhite_c2027_350 111.2 Xgwm124 111.8 Kukri_c21276_1056 113.1 wsnp_Ex_c27176_36393952 114.5 IAAV7428 115.3 BS00063721_51 115.9 wsnp_Ex_c48677_53513032 116.6 RFL_Contig4352_926 118.5 BS00002484_51 119.6 CAP11_c599_115 120.2 Tdurum_contig8580_1285 121.0 BS00077498_51 122.4 BS00035267_51 123.1 Tdurum_contig8840_540 123.5 Excalibur_c12875_1643a 125.8 F109 126.1 Tdurum_contig11046_318 127.2 Excalibur_c98553_61 129.4 AENE-0163_949 130.5 Xwmc44 132.4 BS00015519_51 133.3 Tdurum_contig57153_1439 133.8 Tdurum_contig10475_87 134.9 IACX8074 135.1 IACX2234 135.9 tplb0034g09_1257 136.7 Tdurum_contig29059_185 137.7 Xwmc728 139.4 Tdurum_contig4904_2923 140.2 Xbarc80 141.7 wPt-5253 142.4 wPt-5721 143.1 Xgwm659 144.0 Excalibur_c14706_421 145.6 BS00066052_51 146.5 Wsnp_Ex_c750_1474300 147.1 wsnp_Ex_c11461_18489681 148.7 Tdurum_contig94450_255 152.1 BS00065694_51 153.0 tplb0049h18_765 154.6 RAC875_rep_c111993_160 155.3 Tdurum_contig8669_296 156.0 BS00103710_51a 157.0 RAC875_c14613_112a 158.4 Tdurum_contig13879_352a 159.3 BS00011892_51 160.9 BobWhiBe_c20073_382 161.1 RAC875_rep_c112737_766 162.8 Kukri_c59535_427 164.9 Excalibur_c35345_476 165.2 Tdurum_contig44851_593a 166.7 Excalibur_rep_c102616_317 168.4 Kukri_c94613_316a 169.5 Wsnp_Ex_c750_1474351a 171.3 Tdurum_contig4851_653 172.0 Kukri_rep_c111174_132 173.4 Excalibur_c14806_1091a 174.8 1B T_A, T_2A (CP) TRS,TR V_M C (M acca fer ri2016) MQ TL1 TRL,TR V_CL (M acca fer ri2016) F,C (S M C) TRL (L iu2013) Q RN (C hr ist o p her2013) L,L2,S2,V2 (CP) T2 (S M C) QR D W (K ub o2007) M Q TL2 TRL,ARL_M C (M acca fer ri 2016) Q A rn.1 (G uo2012) SL_CL (M acca fer ri2016) T2 (S M C) M Q TL3 Q R d w .2 (G uo2012) T4 (S M C) Q D RR1 (H am ada2011) LRN_CL (M acca fer ri2016) V (S M C) R T6_M C ( M acca fer ri2016 ) M Q TL4 TRN_M C (M acca fer ri2016) L,L2,L5,S,S1,S2,S5,T ,T1,T2,V ,V1,V2,V5 (CP) F rk,TRL,R V ,S RA (Ib rahim2012) C u lm s (S M C) M Q TL5 QT RSA (B ai2013) LRN _M C (M acca fe rr i2016) M Q TL6 TR V_M C (M acca fer ri2016) TRD ,P RD_M C (M acca fe rr i201 6) SH (S M C) SL_M C (M ac ca fe rr i2016) M Q TL7 TR D_CL (M acca fe rr i2016) R T6_CL (M acca fer ri2016) 8 L T Q M T5_B (CP) SLL,S LSA,S LV (B ai2013) TR N_CL (M acca fer ri2016) L5 ,S5,V5 (CP) M Q TL9 Q M rl .1 (G uo2012) RD W_C L (M acca fe rr i20 16) Q M rl .2 (G uo20 12) MQT L 1 0 TRN,R T6 _CL (M acca fe rr i2016) Q P RE (Ren20 12) MQ TL11 QR V ( Ibr ahim2012) SDW _CL (M acca fe rr i2016) Q R N (Re n2012) M QT L 1 2 T2 (S M C) TRN_ MC ( M ac ca fer ri2016) Figure 2: Continued.
RAC875_c86104_111 0.0 Excalibur_c14401_743 1.9 RFL_Contig4708_1477 2.5 RAC875_c78248_193 4.5 RAC875_c215_329b 6.0 wsnp_Ku_c12399_20037334 6.8 tplb0025f09_1853 8.5 BS00064403_51 11.1 wsnp_Ex_c31121_39955117 12.0 Tdurum_contig49608_1323 15.5 RAC875_rep_c106667_302 18.0 wPt-1708 20.1 Kukri_rep_c117247_191 21.9 Xbarc10b 22.3 wsnp_Ex_c17561_26284693 23.7 wPt-0608 24.4 tplb0050b23_546 25.1 Excalibur_c55463_232 26.0 Xbarc68b 26.5 wsnp_Ex_c29500_38529808 27.4 Xwmc679b 28.1 Xwmc652 29.6 Tdurum_contig29961_68 31.4 GENE-2384_84 32.7 Tdurum_contig97386_207 33.4 tplb0045c06_1675 34.1 Xbarc163 35.2 Tdurum_contig31139_143 35.8 Xwmc310 36.7 Excalibur_c31953_382 37.7 tplb0024a16_411 38.1 Xbarc193 38.8 Xwmc692 39.0 tPt-5519 39.7 Kukri_c4078_3176 40.2 Tdurum_contig74720_539 41.5 Tdurum_contig31514_449 42.9 Tdurum_contig17416_577 43.6 wsnp_Ku_c8075_13785546 44.9 Tdurum_contig93615_735 45.5 Tdurum_contig93615_540 46.2 RAC875_c65971_127 47.3 Xgwm149 47.9 TC67416 48.7 BS00081631_51 49.4 BS00022431_51 50.1 Xwmc617a 51.0 RAC875_c2226_933 52.7 Tdurum_contig28671_295 53.0 Tdurum_contig51688_681 54.3 Tdurum_contig81905_502 54.8 Tdurum_contig42038_2826 55.2 Xgwm1278 55.8 Tdurum_contig42528_936 56.2 Excalibur_c23248_148 56.9 BS00033614_51 57.9 Tdurum_contig33737_277 58.2 Xgwm193a 58.9 BS00065688_51 59.4 Xgwm368 60.4 Xwmc206b 61.0 Xwmc48b 61.4 Xgwm856 61.7 Kukri_c8973_1986 62.2 Tdurum_contig2859_70 62.5 Xgwm513 63.1 Xgwm925 63.5 RAC875_c75075_313 64.0 Xgwm112a 64.7 Tdurum_contig42307_2647 65.6 tplb0043l14_1189 66.6 CA663888 68.0 Tdurum_contig92997_676 68.9 Excalibur_c1043_667 69.4 BS00022177_51b 70.3 tplb0034b12_591 70.9 BS00010940_51 71.5 Xgwm251 71.8 BobWhite_c12067_311 72.5 BS00106144_51 72.9 wsnp_Ra_c3790_6990678 73.5 Xgwm375 74.0 Kukri_c36207_91 74.6 BS00063008_51 74.9 RAC875_c1357_860 75.2 RAC875_c52774_135 75.6 Tdurum_contig29112_483 76.0 wPt-3255 76.7 Tdurum_contig97308_88 77.2 Excalibur_rep_c109675_738 78.0 Kukri_c6388_1030 78.4 Tdurum_contig10466_78 79.0 Tdurum_contig45927_677 79.4 Excalibur_c27139_396 79.7 Tdurum_contig82603_67 80.6 tplb0036a20_1295 80.9 Tdurum_contig50783_285 81.3 BS00023035_51 81.7 RAC875_c36213_352 83.1 Tdurum_contig46788_529 83.5 Excalibur_c49297_159a 84.0 tplb0056i06_856 84.8 CAP12_c4704_232 85.0 Tdurum_contig920_126 85.8 Ex_c45493_761 86.6 wsnp_Ex_c5187_9195120 87.0 Tdurum_contig76671_435a 88.5 BobWhite_c8115_648 89.3 Tdurum_contig60051_169a 89.6 Kukri_rep_c103450_1504 90.1 Kukri_c2639_290 90.6 wsnp_Ex_c38704_46166494 91.2 GENE-2826_325 92.1 Kukri_c44559_429 92.7 BS00004727_51 94.0 Tdurum_contig9893_571 95.0 Kukri_rep_c78644_408 95.6 Tdurum_contig51521_90 96.1 Kukri_c93922_68 97.0 Kukri_c93922_206 97.7 wPt-0391 98.3 IAAV5564 98.7 BobWhite_c27801_429 100.3 BobWhite_c4818_173 101.3 D_F5XZDLF01C3B2R_358 102.3 Ra_c19259_1799 103.1 Excalibur_c29255_366 103.7 wsnp_BG604404B_Ta_2_1 104.0 BS00022366_51 104.8 RAC875_c10029_341 105.0 IAAV7394 105.7 IAAV8499 108.2 Tdurum_contig56887_501 108.7 wPt-5564 108.8 Tdurum_contig56887_322 109.1 BobWhite_c11977_428 109.2 BobWhite_c47144_153 109.5 wPt-8292 110.3 wPt-3804 wPt-5996 110.5 Tdurum_contig52336_160 110.8 Tdurum_contig56887_256 111.6 RAC875_rep_c117991_66 112.0 JG_c3271_440 112.1 Tdurum_contig9168_2214 112.7 BS00034148_51 113.7 BS00034147_51 114.3 BobWhite_c15529_288 114.9 RAC875_rep_c82932_428 115.5 RAC875_c17026_714 116.5 JD_c48945_490 116.7 IAAV3549 117.4 Xdupw43 117.7 BS00004012_51 117.9 BS00065222_51 118.8 BS00014101_51 118.9 Tdurum_contig42695_410 119.3 RFL_Contig3368_209 119.4 Tdurum_contig9118_1078 120.9 RFL_ConBiA3368_209 121.3 RAC875_c6026_963 122.9 Kukri_rep_c86966_239 123.2 4B Kukri_c3009_1822 0.0 BS00003772_51 2.9 RAC875_c16610_470 3.8 RAC875_c13610_822 4.7 GENE-1756_564 6.4 Tdurum_contig55193_347 7.0 Xpsr899 10.6 BobWhite_c704_596 11.9 Kukri_c3827_337b 12.1 Tdurum_contig12045_327 13.5 Ku_c17678_1161 14.9 RFL_Contig5170_1904 15.7 BS00077635_51 16.0 wPt-1377 17.0 wPt-1664 18.1 BS00082191_51 19.3 wPt-9382 20.4 RAC875_c106584_747 21.1 wPt-0562a 21.4 BobWhite_c15802_720 22.2 Kukri_c49017_404 23.2 wPt-9679 24.0 wPt-7565 24.9 wPt-2153 25.5 wPt-0495 26.2 wPt-8266 27.0 BobWhite_c5092_422 27.5 BS00022032_51a 28.0 tPt-0877 28.5 Kukri_c34219_355 29.0 wPt-3468 29.5 Xgwm334 30.2 Ex_c68796_2057 30.8 wsnp_Ku_c20866_30535489 31.4 Excalibur_rep_c68175_305 33.1 Xgwm1040 34.7 Excalibur_c5442_1691 35.7 wPt-7663 36.9 Ku_c10377_335 37.6 BS00059475_51b 38.1 Excalibur_rep_c102994_1689 39.8 RAC875_c21158_99a 40.1 BS00110963_51 42.8 tplb0039m17_2125 43.3 wPt-2822 44.5 BobWhite_c36070_89 45.2 wsnp_Ex_c19873_28889970 45.6 wPt-9692 45.8 RAC875_c38494_52 46.6 BS00065496_51 47.4 BobWhite_c13839_111 47.7 wsnp_Ex_c1011_1931797 48.3 wPt-7623 48.6 Excalibur_c62661_195 49.0 RAC875_c53520_103 49.7 Xgwm825 50.2 Excalibur_c14693_724b 51.5 Excalibur_c14222_179 52.2 wsnp_Ex_c31149_39975724 52.7 BS00066452_51 53.0 Xbarc23a 53.6 tplb0048b19_119 54.1 IAAV4117 55.1 wsnp_Ex_c31149_39976066 55.5 wPt-9829 55.9 wPt-10645 56.7 RAC875_c5395_564 57.2 BS00063990_51 57.9 BS00023627_51 58.7 Jagger_c2853_75 59.2 Tdurum_contig50698_601 59.7 wsnp_BE490604A_Ta_2_1 60.1 Xpsr312b 60.9 wsnp_Ex_c14692_22766127 61.2 CAP7_c11100_94 62.2 Ku_c11950_931 62.5 IAAV5188 63.1 RAC875_c64560_111a 64.2 TA004097-0977 64.8 Kukri_c16868_1062 65.0 Xgwm82 65.5 Xwmc243b 65.6 BS00036878_51 66.0 Xgwm356b 66.4 Xwmc807 67.0 BS00064548_51 67.7 wsnp_Ex_c341_667884 67.9 Xwmc335a 68.6 Xwmc684 69.4 Tdurum_contig43974_219 69.6 Xpsr142 70.0 Xgwm907b 70.4 Excalibur_c2287_1327 71.5 Xbarc113 72.7 tPt-4209 73.0 Xwmc553 74.0 wPt-7063 74.5 Xwmc179b 76.0 Xwmc163 76.5 wsnp_Ra_c12086_19452422 76.8 BobWhite_c13845_195 77.4 BS00040166_51 78.6 Tdurum_contig82427_252 79.4 Kukri_c4458_1060 80.0 Tdurum_contig75814_655 80.7 Excalibur_c8784_1005 81.5 wsnp_Ex_c26147_35395259 82.0 Tdurum_contig54796_1427 84.0 CAP7_rep_c11162_75 84.4 Excalibur_c25898_434 85.0 CAP11_rep_c6864_291a 85.4 Kukri_c9763_115 85.8 wsnp_Hu_c1075_2160065 86.0 BobWhite_c14304_687b 86.3 BS00065700_51 86.6 Tdurum_contig7464_1168 89.2 Excalibur_c5868_1486 89.6 Ku_c604_705 90.0 wsnp_Ex_rep_c68010_66754171 90.7 Ex_c5868_1113 91.1 CAP7_c9578_263 91.9 IAAV3609 93.6 BQ838884 95.7 Tdurum_contig97355_136 96.6 Tdurum_contig62188_108 97.0 TA001855-0472 98.1 Tdurum_contig14676_637 100.2 Tdurum_contig92441_354 102.0 Xgwm169 102.9 Tdurum_contig43566_822 103.9 Tdurum_contig29357_338 104.3 Tdurum_contig43566_801 105.2 Kukri_rep_c70063_663 105.9 wPt-11612 106.6 Xwmc417a 107.4 wsnp_Ra_c5346_9501281 108.0 wsnp_Ex_c8510_14306239 108.8 wsnp_Ex_c20457_29526403 110.0 Xcdo1091a 110.8 RAC875_c103443_475 111.9 Tdurum_contig28802_213 112.4 Xwmc580 112.8 Kukri_rep_c95718_868 113.4 wPt-8373 113.9 wPt-11668 114.3 wPt-2877 114.8 Xgwm427 115.1 BobWhite_c232_563 115.6 tPt-6661 116.8 Xgwm617b 117.2 wsnp_Ku_c1318_2624758 117.4 BobWhite_c5872_589 118.3 RAC875_c2090_219 118.8 wsnp_JD_c26552_21868492 119.2 wPt-7390 120.1 wPt-1375 120.8 Xcdo836 121.7 Excalibur_rep_c103232_355 122.2 Xpsr546a 122.9 RAC875_c26158_376 123.2 RAC875_c10648_86a 123.9 RFL_Contig2765_1148a 124.2 Tdurum_contig51627_1211 125.1 Excalibur_c79991_552 125.5 Tdurum_contig43308_465 125.7 F133 126.0 wPt-2975 126.6 wPt-9976 127.1 wPt-2632 128.3 BS00062776_51a 129.1 Xbarc28b 129.8 Excalibur_c64804_348 130.5 wPt-6829 131.0 D_contig37522_188a 131.6 Xgwm999 132.8 Tdurum_contig27971_183a 133.2 CAP8_c950_198a 134.9 Ra_c74397_248 136.7 BS00064970_51a 138.9 Xwmc621 141.2 BS00086174_51 143.3 Tdurum_contig17378_299 147.5 Excalibur_c35871_596 148.2 GENE-3993_694a 149.1 wsnp_JG_c5646_2148296a 150.4 BobWhite_s67399_107 152.0 TA005679-0546b 158.5 Kukri_c67474_466a 162.2 BS00064558_51a 163.1 Tdurum_contig10729_118a 165.7 Ex_c10413_1606a 167.7 Excalibur_c37062_65a 168.6 Excalibur_c62474_82 170.9 Swes123 172.4 Xwmc254c 181.5 6A tplb0060p09_735 0.0 Kukri_c14683_65b 2.3 Xwmc728b 3.1 BobWhite_rep_c63710_181 3.9 Xgwm234 4.4 wPt-1302 5.1 wPt-9800b 8.0 wPt-0033 10.0 Xgwm443b 12.2 wsnp_Ex_c33932_42333941 13.6 RAC875_c103396_446 14.6 RAC875_rep_c106241_304b 15.6 wPt-0819 17.6 Tdurum_contig9291_553b 18.1 BS00064042_51 18.7 wPt-8604 19.7 wsnp_Ex_rep_c110196_92676847 20.2 wsnp_Ku_c5308_9450093 21.4 BS00015136_51 22.6 RAC875_c18322_670 23.4 Excalibur_c40676_447a 25.0 Excalibur_c3730_546 26.5 wsnp_Hu_c568_1187615 27.4 Ku_c3398_941 28.3 wsnp_Ku_c35090_44349446 29.6 Xwmc149b 30.1 Ra_c19173_1010 32.1 Tdurum_contig16267_354 33.0 Kukri_rep_c106411_137 34.3 Xwmc274b 34.8 BS00063769_51 35.0 Xgwm544a 35.4 Xgwm66c 36.2 BobWhite_c43220_84 37.0 RAC875_c60836_741a 38.3 JD_c4730_663 38.9 wsnp_Ex_c2571_4784380 39.4 wsnp_Ex_c13468_21202450 40.4 wsnp_RFL_Contig4574_5416228 41.3 wPt-5914 42.1 wPt-6348 43.1 Xwmc386 43.5 wsnp_Ex_rep_c68119_66884852 45.4 RAC875_c4219_1340 46.5 wsnp_BE497820B_Ta_2_3 47.0 Xbarc216 48.2 tPt-0228 48.9 IAAV1835 49.4 Xgwm335 50.4 Kukri_c22967_1272 51.0 Excalibur_c8082_478 51.5 Xwmc616 52.1 Xbarc74 53.5 BS00064578_51 53.9 Xbarc89 54.1 Xbarc176a 55.1 Tdurum_contig42092_947 55.5 Xgwm564b 56.0 BS00031411_51 56.6 Xgwm213a 57.3 GENE-3211_372 58.7 Xwmc435a 59.3 wsnp_Ex_c974_1864971 60.0 IAAV5974 60.3 wsnp_Ra_c15297_23684845 60.6 wsnp_Ra_c26091_35652620 61.7 Tdurum_contig10172_176 62.5 Xwmc745 63.2 Kukri_rep_c69515_183 64.3 Xgwm777 64.6 wPt-0935 65.0 Xgwm371 67.1 wsnp_Ex_c2727_5053747 68.3 Xwmc415a 68.6 wsnp_Ex_c28687_37791888 69.5 RFL_Contig2304_1569 70.8 BS00015205_51 71.1 BobWhite_c6633_240 71.9 wPt-4936 72.3 Xgwm639b 73.9 wPt-7167 74.0 Xwmc405a 75.6 Kukri_c3541_185 76.9 Xwmc759 77.3 wPt-3661 78.0 IAAV2509 78.4 Kukri_c6424_810 79.0 wPt-3457 81.2 wPt-1250a 82.8 wPt-3503a 83.6 Kukri_rep_c102132_410b 85.6 wPt-9613 87.5 Kukri_c1137_1421 88.7 Kukri_c18158_354 89.5 BS00009335_51 90.9 Excalibur_c80450_75b 91.2 Xwmc537a 92.4 Xpsr911b 93.0 BS00022582_51b 94.2 wPt-6135 95.1 Tdurum_contig97342_274 96.6 wsnp_Ex_c13485_21225504 97.3 Tdurum_contig29319_256 98.0 Xgwm554a 99.3 Tdurum_contig44181_730 99.8 Tdurum_contig27797_654 101.0 Ra_c69621_237 103.2 BS00009443_51 104.2 RAC875_rep_c102342_470 105.5 Excalibur_c32708_225 106.6 IACX5702 107.2 RFL_Contig5461_683 108.3 wPt-9454 109.6 Excalibur_c5329_1335 110.2 wsnp_Ex_rep_c68600_67448893 111.0 RFL_Contig539_1789 112.4 Kukri_c59540_137 114.5 tplb0053c01_1628 115.7 Kukri_c2955_281 116.1 Kukri_c57954_369 117.2 Ra_c2216_1442 118.0 Excalibur_c100531_251 119.0 Excalibur_rep_c113786_110 119.5 Tdurum_contig29967_456 120.9 BS00064593_51 121.2 Tdurum_contig10793_127 122.0 Tdurum_contig47491_258 123.0 wsnp_Ex_c12152_19431363 124.6 Tdurum_contig70554_1006 125.1 wPt-6989 126.1 Xwmc289a 128.0 Kukri_c10508_253 129.0 wPt-1482b 130.1 tPt-1253 130.9 Xwmc500f 131.7 Tdurum_contig42069_208 132.7 Xwmc160 133.6 Xbarc232b 134.9 Xwmc326d 135.8 Kukri_rep_c113552_119 136.8 Tdurum_contig97407_196 137.1 BS00037023_51 138.7 Xcfd86b 139.2 Xwmc28 140.9 Tdurum_contig32091_165 141.7 Xwmc235 142.9 BS00012140_51 143.2 Excalibur_c17489_804 144.9 Xwmc118 145.2 Ra_c6374_1984 146.1 Xwmc508 147.5 Tdurum_contig58293_755 148.6 Excalibur_c53149_153 149.2 Excalibur_c4699_147 150.2 Kukri_c27433_2333 151.3 BS00062618_51 152.1 GENE-4868_140 153.0 wsnp_Ra_c38873_46699852 154.1 RAC875_c88582_131b 157.1 Tdurum_contig81337_335 158.6 wsnp_Ex_c2870_5296539 159.6 IACX751 160.3 wPt-8418a 161.2 wPt-7561 162.2 Xwmc640b 164.5 Excalibur_c9543_1268 165.4 BS00069417_51 167.3 wsnp_Ku_c38713_47298856 168.1 BS00033182_51 170.5 Tdurum_contig45588_730 171.4 Gs-41a 172.8 wsnp_JD_rep_c50403_34392266 174.0 wPt-1348 175.6 wPt-1179 176.8 Kukri_c1214_2316 177.6 Xwmc430e 178.1 Xbarc59a 178.9 Excalibur_c2618_1412 179.3 BobWhite_c7818_278 179.6 BS00022876_51a 180.1 BS00031338_51 181.1 CAP11_c2765_78 182.1 wPt-0566 183.2 Tdurum_contig13965_1223 183.9 BAC875_Bep_c106941_417 184.3 Xwmc783 185.3 Tdurum_contig5522_455 186.2 BobWhite_c15406_510 188.3 BS00024829_51 189.5 Y4c 190.5 TC88560 194.5 Tdurum_contig51669_668 198.3 RAC875_c14078_202a 199.8 Xgwm497d 200.8 Tdurum_contig13489_292b 202.6 Excalibur_c9969_98 204.0 Kukri_c84307_76 205.3 Excalibur_c34579_263 206.1 wPt-7665 208.8 Tdurum_contig42257_3252 209.6 Kukri_c7546_1739 211.5 wsnp_Ex_c24031_33277293 212.4 wPt-9116 213.3 Kukri_c5318_380 215.7 Tdurum_contig43552_666a 217.0 wPt-1500 217.3 wPt-5373 218.9 wPt-5168 219.7 TC86533 222.0 wPt-0484 223.9 5B Rht-1B TRD_M C (M acca fer ri2016) SD W_M C (M ac ca fe rr i2016) SL_M C (M acca fer ri2016) M Q TL13 LRN_M C (M acca fer ri2016) V ,L,SA,T ,S H,LD W ,RD W ,F ,C (S M C) LRN_M C (M acca fer ri2016) M Q TL14 LRL,TRL ,RN (Ren20 12) PR V_M C (M acca fer ri20 16) RD W ,RS R_CL (M acca fer ri2016) M Q TL15 R T6_CL (M acca fer ri2016) R GA_CL (M acca fer ri2016) M Q TL16 TRN_M C (M acca fer ri2016) RG A _M C (M acca fer ri2016) TRS_M C (M acca fer ri2016) TRN _CL (M acca fer ri2016) PRL,P RS_C L (M acca fe rr i20 16) R T6_CL (M ac ca fe rr i2016) TR L_M C (M acca fe rr i2016) F,SA2,V2, L2 (S M C) M Q TL17 TR V_M C (M acca fer ri2016) L,L1,SA,SA1,V1 (S M C) M Q TL18 ARL_M C (M ac ca fer ri2016) R GA_M C (M acca fe rr i2016) M Q TL19 Q RN (Zha n g20 13) SD W_M C (M acca fer ri20 16) MQT L 20 T TRL,S LL,S LSA,S LV (B ai2013) SL_M C (M ac ca fe rr i2016) TRL,PRA,RSA (L iu2013) MQT L 2 1 T7 ( SMC ) RD W_M C (M acca fer ri2 016) Q RN (Ch ri st o p her2013) MQT L 22 TR L (Ren2012) RD W (Y u2013) RD W (G uo2012) TRL (B ai2013) L,L1,L2,S,S1,S2,V1,V2,T ,T1 (CP) MQ TL23 SL V (B ai2013) RT 6 _M C (M acca fer ri2016) MQ TL24 SAL (B ai2013) SL_CL (M acca fer ri2016) M Q TL25 LRL (Ren2012) RD W (G uo2012) SH ,L DW ( SM C ) MQT L 2 6 TRL_M C (M acca fer ri2016) Q P VRN (K ub o2007) MQT L 2 7 TRL ,R GA,ARL_CL ( M acca fer ri2016) T, L 1 ,SA1,T1 (S M C) PRS_ M C (M acca fe rr i2016) TRS,PR V_M C (M acca fe rr i2016) PRL _ MC ( M ac cafe rr i2016) MQT L 2 8 RG A _ MC ( M ac ca fer ri2016) RD W (G uo2012) Figure 2: Continued.
RFL_Contig6075_1128 0.0 BobWhite_c20735_255 3.0 Tdurum_contig11531_522 8.6 Xgwm255 10.6 wPt-0276 11.8 Jagger_c9100_76 12.6 Xwmc606a 13.7 Xwmc323 14.8 wPt-3147 16.2 Xgwm569 18.3 Tdurum_contig16244_335 19.2 Excalibur_c50044_1221 21.9 Kukri_c50501_299b 22.4 Tdurum_contig100306_98 23.0 BS00062990_51 24.5 Tdurum_contig30677_55 27.3 BJ239878 28.5 wsnp_CAP8_c334_304253 29.5 RAC875_c23521_589 31.9 wPt-2278 32.5 Xwmc338 34.7 wPt-4309 35.4 BobWhite_c41356_62 36.1 Kukri_c67849_109 38.2 Xwmc597f 39.0 BS00081132_51 40.3 Excalibur_c29698_76 42.4 IAAV5530 43.7 BobWhite_c44404_312 44.3 Kukri_c109962_396 46.2 Excalibur_c108159_334 47.6 wPt-2883 48.6 Excalibur_c41298_459 49.4 wsnp_Ex_c2103_3947695a 50.4 Xgwm46b 51.8 Xgwm601a 52.9 wsnp_Ex_c23755_32994701 53.8 Xgwm951 55.0 GENE-0293_65 57.1 wPt-8283 58.3 Xgwm68b 59.8 RAC875_c5683_136 62.5 Xwmc546a 63.3 Tdurum_contig68347_605 64.6 RAC875_c1638_165 65.4 RAC875_c56878_518 67.0 Xbarc85 68.9 Xpsr103 69.6 wPt-4230 70.5 wPt-9630 71.4 wsnp_Ex_c10251_16815792b 71.7 Xgwm963b 72.2 Xbarc65 73.4 Xwmc653b 74.7 Xgwm213b 75.5 wPt-3873 76.1 Kukri_c2796_1436 77.0 TC69176 78.0 Kukri_rep_c69312_647 78.7 wPt-3833 79.1 BobWhite_c27679_112 79.8 BobWhite_rep_c64772_309 80.2 wPt-4025 80.9 wPt-0920 81.3 BobWhite_c3541_152 82.0 wPt-8919 82.9 Kukri_rep_c71173_2043 83.8 RAC875_c28901_836 84.5 Tdurum_contig9966_724 85.3 Tdurum_contig9966_646 86.1 Xgwm1036 86.6 wPt-6180 87.1 wPt-4323 88.2 Xwmc723 88.6 Tdurum_contig49861_368 89.1 wPt-8069 89.9 Xdupw403 90.8 wsnp_Ex_c758_1488368 91.0 Xdupw145 91.6 BS00063208_51 92.0 Excalibur_c11857_1196 92.4 RAC875_c21537_368 93.2 BS00029286_51 94.8 tplb0033d17_389 95.7 Xbarc1181 98.0 Tdurum_contig81911_179 98.7 Excalibur_c33549_95 99.3 wPt-2273 99.7 Xwmc517 100.3 tplb0062h23_1362 101.2 Excalibur_c93217_61 102.4 tplb0021f14_1700 103.2 Xwmc537b 104.8 BS00080621_51 105.4 Xbarc272 106.6 Tdurum_contig14821_751 107.5 Excalibur_c33267_263 109.1 BS00064871_51 111.1 Kukri_c15912_1189 112.4 BS00009995_51 113.9 Ku_c31231_285 114.8 wPt-5922 115.2 wPt-1826 116.5 wPt-4258 117.0 RFL_Contig4842_579 117.6 wPt-7887 118.0 Excalibur_rep_c74778_252 118.7 Xwmc792b 120.1 Excalibur_rep_c104090_439 120.8 RAC875_c41903_122 121.6 Xdupw398 122.7 wPt-5341 123.9 tPt-7362 126.1 Xwmc311 127.3 rPt-3887 128.6 wPt-5343 129.0 wsnp_Ex_rep_c68762_67626384 129.8 Excalibur_c12644_58b 130.7 Kukri_c10430_1138 131.1 Xgwm420 132.0 BobWhite_c12048_145 133.2 IAAV9045 134.1 wPt-4297 135.0 wsnp_Ex_c323_629461 136.0 Excalibur_c19471_197b 137.3 wPt-2356 138.5 wPt-4814 139.1 RAC875_c31791_559 140.1 BobWhite_c26534_532 140.9 Ra_c3092_741 142.7 Xgwm611 143.5 wPt-0600 144.4 Tdurum_contig8402_638 144.8 wPt-3939 145.0 wPt-5138 146.2 RAC875_c49954_1172 147.0 Xgwm577 147.5 Xbarc1073 148.5 wPt-7219b 149.2 Excalibur_s103240_100 149.9 wPt-2677 150.2 RAC875_c10372_671 150.6 Tdurum_contig11832_272 151.2 Xwmc500b 151.8 Tdurum_contig28598_245 152.4 wPt-4902 152.9 Kukri_c7284_1859 153.4 Xbarc182 154.0 BS00066342_51 154.8 Xcfa2040c 155.2 wPt-9877 156.4 Xbarc32 157.2 Xmag1933 158.1 wsnp_BE445506B_Ta_2_1 159.4 wPt-7891 160.2 Xwmc557 160.8 wPt-0504 161.9 Tdurum_contig45280_451 162.2 wsnp_Ex_c12556_19991329 163.0 Xwmc10 163.8 CAP7_c12398_110 164.2 tplb0040b02_465 165.0 Excalibur_rep_c110429_536 166.2 Xwmc526 166.8 RAC875_c525_941 167.3 Tdurum_contig30909_76 168.0 wPt-8650c 168.7 Xwmc70 169.8 wPt-6865 170.6 RAC875_c9543_99b 171.6 wPt-0465 172.9 wPt-1069 174.1 Xgwm344c 175.7 wPt-7387 176.1 wsnp_Ex_c53387_56639949b 177.1 Ex_c29172_877b 178.9 Tdurum_contig13459_543a 180.7 Excalibur_c1750_188 181.9 GENE-3073_258 183.1 RAC875_c1329_488 188.6 wPt-9488b 197.3 7B BS00016587_51b 0.0 Xwmc430d 1.0 Excalibur_c28854_1580 2.0 BS00093063_51a 2.6 Xgwm1051 3.7 wsnp_Ex_rep_c101133_86572194 4.0 BS00029434_51 5.3 Tdurum_contig43538_1582 6.0 wPt-8641 6.5 Xdupw217 6.9 wPt-0151 7.0 RAC875_c841_1329 8.1 wPt-4720 8.4 wPt-2714 9.2 wPt-3304 10.0 wPt-7777 11.0 tPt-0910 11.8 RAC875_c63707_140 12.3 wPt-6988 13.4 wPt-1474 14.0 wPt-8015 15.7 RAC875_c12259_1892 17.4 RFL_Contig5885_435 18.3 Kukri_rep_c111561_377 18.9 wPt-4233 19.8 wPt-4706 20.9 Tdurum_contig10149_284 21.3 Kukri_c67545_299 22.2 wPt-7954 23.2 wPt-1437 23.9 Kukri_c41439_313 24.3 wsnp_Ra_c30621_39857295 25.5 GENE-1970_158b 26.4 wsnp_Ex_c14481_22485922 27.2 wPt-2095 27.8 BS00033629_51 29.6 GENE-1999_98 30.4 wPt-0259b 31.0 BS00035056_51 32.0 Xwmc419c 33.0 wPt-3376 33.5 Xwmc486 33.9 wsnp_Ku_c15011_23496657 34.5 Tdurum_contig50121_249 35.0 Kukri_c60727_356 35.5 wPt-5256 35.9 wPt-1241 36.8 BS00067133_51 37.1 wPt-3605b 38.0 wPt-2786 38.9 wPt-2175 39.8 Xwmc487 40.5 Kukri_c46237_140 40.8 wPt-4858 41.3 TA004917-1090 42.0 Tdurum_contig11641_1008 42.8 Xwmc597d 43.1 Xgwm508 43.8 wPt-2479 44.0 wPt-0470 44.5 JD_c63404_542 44.7 wPt-5333 45.1 wPt-3163 45.4 Excalibur_c55800_202 46.4 wPt-2218 46.8 Kukri_c21995_1652 47.0 wPt-8721 48.1 TC85037 48.7 wPt-7489 49.6 RAC875_rep_c111705_629 50.2 XksuD17 50.5 Xwmc494 51.0 IAAV8375 52.0 GENE-4142_601 52.9 Xgwm518 53.4 RAC875_c23812_187 53.7 Xbarc68c 54.0 Xgdm113 54.3 BobWhite_rep_c65142_111 57.1 CAP8_rep_c9477_231 58.1 wsnp_Ex_c6057_10611952 59.0 Xbarc101 59.9 wsnp_CAP7_c1839_907899b 60.3 Xbarc1169 60.8 BS00046966_51 61.7 Tdurum_contig62040_1253 62.0 wsnp_BE488206B_Ta_2_1 62.5 Ra_c19002_569 63.0 IAAV7767 63.8 wsnp_Ex_c18382_27210656 64.2 Tdurum_contig21737_203 64.7 RAC875_c20889_913 65.1 Xwmc398b 65.7 RAC875_c30027_381 66.3 Xwmc265c 67.2 Xgwm193b 68.7 wPt-0446 69.0 BobWhite_c686_387 69.4 wsnp_Ex_c5058_8981554 70.2 Kukri_c25589_772 71.2 BS00067644_51 71.8 Excalibur_rep_c101274_113 72.1 wsnp_Ra_c11942_19247669 73.0 tplb0059j12_800 74.0 wPt-9124 74.5 Xgwm88 74.9 Xgwm644 75.3 Xbarc198 76.0 Xgwm887 76.7 Tdurum_contig69600_415 77.2 Kukri_c45250_289 77.8 wPt-0397 78.7 Excalibur_rep_c94584_98 79.1 Tdurum_contig93283_513 79.7 Xgpw3241 80.2 wsnp_Ku_c16522_25425455 80.6 wPt-9241 81.1 TA005272-0214 81.3 Xwmc152 81.6 Tdurum_contig47269_241 82.1 wsnp_CAP11_c1432_806102 82.9 Xbarc79 83.1 wPt-8554 83.6 wPt-1730 83.9 CAP7_c7065_166 84.6 wPt-1287 85.2 RAC875_c6649_642 85.7 BobWhite_c30500_527a 86.2 Xwmc539 86.7 Excalibur_c11817_1125 87.1 Xdupw216 87.7 GENE-4221_313 89.1 BobWhite_rep_c53133_385 90.1 BS00037784_51 92.4 Xbarc178 92.9 Kukri_c49331_77 93.0 Xgwm219 93.7 RAC875_c4891_99 95.0 wPt-9930 95.7 wsnp_Ex_c18632_27501724 96.7 Xwmc417b 98.4 wsnp_Ku_c11846_19263340 99.2 tplb0021a17_853 100.2 RFL_Contig2785_708 100.8 Kukri_c5168_162 101.2 BS00067129_51 101.6 Xgwm768 102.0 BS00063595_51 103.3 Kukri_c40428_72 104.7 wPt-7830a 105.1 tPt-9948a 106.0 wPt-6184 106.6 Excalibur_c47675_176 107.2 wPt-5092 107.5 wsnp_RFL_Contig3708_3946935a 107.9 BobWhite_c3514_717 108.7 Tdurum_contig71517_158 109.2 wPt-7274 109.7 wPt-9481 110.8 wPt-2178 111.6 Xgwm735 112.2 RAC875_c16691_90 113.1 BS00068815_51 114.8 IACX2322 115.9 wPt-9952 116.2 RAC875_c3455_171 117.0 wPt-1325 118.4 BS00088277_51 119.4 JD_c7044_1403 121.9 BS00068166_51 123.0 RFL_Contig6086_1186b 123.6 Excalibur_rep_c105053_313 124.6 BS00049082_51b 125.1 RAC875_c17224_741 125.8 IAAV5595 126.3 GENE-3988_631 126.7 BobWhite_rep_c63427_478 127.0 Excalibur_c27139_516 128.2 Tdurum_contig7986_704 128.6 tplb0046e21_69 129.2 RAC875_c22494_231 130.0 Xwmc388 130.9 wsnp_Ex_c293_567035 131.4 RAC875_c55445_92 132.2 BobWhite_c27364_124 133.4 Excalibur_c14451_1137 133.9 wsnp_Ex_c791_1557333 134.6 wsnp_CAP7_c1735_859744 135.0 Excalibur_rep_c69865_157 135.9 BS00022278_51 136.6 RAC875_rep_c108385_407a 137.1 IACX7844 138.7 BobWhite_c10171_323a 138.8 Excalibur_c6108_262 139.0 TA002907-0816 139.3 Kukri_c36312_591 139.8 tplb0045b09_1555 143.9 Tdurum_contig42191_4367 144.4 wPt-6329 157.7 wPt-0171 161.5 wPt-5885 162.8 wPt-5480 166.4 wPt-5176 167.9 wPt-1541 170.4 6B LRN_M C (M acca fer ri2016) R T6,ARL_M C (M acca fer ri2016) T ,T1 (S M C) MQT L29 Q RN (Ren2012) Q LRL (Ren2012) Q MRL (G uo2 012) SL_CL (M acca fer ri2016) PRS,PRL_M C (M acca fer ri2016) TR D_CL (M acca fe rr i2016) M Q TL30 P RL,TRL _M C (M acca fer ri2016) TL4,L5,L6,V4,V5,V6,SA4,SA5,SA6 (S M C) T_A (CP) MQT L 3 1 Q MRL (Ren2012) MRL,PRE (Ren2012) L9,S9 ,SA9 (S M C) SDW _ CL (M acca fe rr i2016) R T6_M C (M acca fe rr i2 016) M Q TL32 PRL (Ren2012 ) MRL (Ren201 2) M Q TL33 QM RL (L iu201 3) PRS,P RL_M C (M acca fer ri2016 ) TRS,TRL_CL (M acca fe rr i201 6) ARL_CL (M acca fe rr i2016) MQT L 3 4
Figure 2: Integrated map in wheat for the QTL and the MQTL identified through the meta-analysis for the root morphological traits. QTL from literature are not indicated with the QTL name reported in the correspondent study but with a nomenclature in which the trait and the reference are reported, as indicated in Table S1, for clarity. The same for the QTL identified in the present study, which are reported with the
acronym“SMC” (i.e., “Simeto” × “Molise Colli” population). The genetic position of the Rht-B1 locus on chromosome 4B is also reported.
Vertical lines on the right of the chromosomes indicate the confidence intervals, and horizontal lines indicate the peak marker positions,
where the length represents the percentage of variability explained by the QTL. The MQTL are in bold, while the single QTL are in gray. The names of the QTL grouped in the same MQTL are in the same color.