Review
P53
and
Sirt1:
Routes
of
metabolism
and
genome
stability
Stefania
Gonfloni
a,b,*
,
Valentina
Iannizzotto
a,1,
Emiliano
Maiani
a,1,2,
Giovanna
Bellusci
a,
Sarah
Ciccone
a,
Marc
Diederich
ca
DepartmentofBiology,UniversityofRome‘‘TorVergata’’,ViadellaRicercaScientifica,00133Rome,Italy
b
LaboratoiredeBiologieMole´culaireetCellulaireduCancer,KirchbergHospital,9RueEdwardSteichen,2540Luxembourg,Luxembourg
c
CollegeofPharmacy,SeoulNationalUniversity,1Gwanak-ro,Gwanak-gu,Seoul151-742,RepublicofKorea
Contents
1. P53,effectonmetabolismandoxidativestress... 000
1.1. TheWarburgeffectandp53 ... 000
1.2. P53andglucosemetabolism... 000
1.3. P53modulatestheglucosemetabolismwhilecontrollingthegluconeogenesis... 000
1.4. P53andoxidativephosphorylation... 000
1.5. P53lipidmetabolismandmitochondrialhomeostasis... 000
2. EssentialeffectsofSIRT1onmetabolismandgenomeprotection... 000
2.1. Metabolismandsignaling ... 000
2.2. Genomicstability... 000
2.3. DNArepair ... 000
3. SIRT1-p53:nodesconnectingmetabolismandgenomeintegrity... 000
4. Conclusionsandperspectives... 000
Acknowledgements... 000
References... 000
1. P53,effectonmetabolismandoxidativestress
Metabolicchangestakeplacewithtumorprogression.Cancer cells often rewire their metabolic pathways to promote fast growingandgenomicinstability.Thebest-understoodfunctionof p53 is itscentral role astumor suppressor. Emergingevidence indicatesaroleofp53inmonitoring/modulatingcellmetabolism
[1].Cellfatedependseitherbyp53transcription-dependentor -independentresponseswithinmitochondria.P53regulatesmany
ARTICLE INFO
Articlehistory: Received28June2014
Receivedinrevisedform28August2014 Accepted29August2014 Availableonlinexxx Keywords: p53 SIRT1 Metabolism Tumorsuppression Genomeintegrity ABSTRACT
Thetumorsuppressorp53isatranscriptionfactorthatregulateskeyprocesses.But,theoutcomesofthe p53responsegobeyonditsroleasanucleartranscriptionfactor.Sirtuin(SIRT1)regulatesp53functions astranscriptionfactor.Atthesametime,SIRT1protectsthegenomeunderstressconditions.Thelink betweenp53andSIRT1responsesisunique.Bothregulatemetabolism,stresssignaling,cellsurvival,cell cyclecontrolandgenomestability.Recentstudieshaveproposedcancerasametabolicdisease.Thisis duetotheswitchfromaerobictoanaerobicmetabolismduringtumordevelopment.Yet,thecomplex molecularcircuits(inandoutofthenucleus)oftumorprogressionremainelusive.Inthisreview,wewill focus onthe interplaybetween p53 andSIRT1. We willdiscuss theirroles asnodes forpossible therapeuticintervention.
ß2014ElsevierInc.Allrightsreserved.
* Correspondingauthor.
E-mailaddresses: Stefania.Gonfl[email protected], Stefania.Gonfl[email protected] (S.Gonfloni).
1Theseauthorscontributeequallytothiswork.
2Presentaddress:CellStressandSurvivalUnit,DanishCancerSocietyResearch
Center,Strandboulevarden49,2100Copenhagen,Denmark.
ContentslistsavailableatScienceDirect
Biochemical
Pharmacology
j our na l ho me p a ge : w ww . e l se v i e r . com / l oc a te / b i och e mph a rm
http://dx.doi.org/10.1016/j.bcp.2014.08.034 0006-2952/ß2014ElsevierInc.Allrightsreserved.
proteins required for metabolism and reactive oxygen species (ROS)production.Besides,p53controlsredoxsignaling,through themodulatedexpressionofpro-andanti-oxidantproteins[2].
1.1. TheWarburgeffectandp53
Transformedcellsgetaseriesoffeaturestoproliferatefastand toescape from programmedcell death. In 1927 Otto Warburg demonstratedthattumorprogressiontakesplacetogetherwith metabolicchanges[3].Warburgobservedthat cancercells shift from oxidative phosphorylation (OXPHOS) to glycolysis. This occurseveninpresenceofoxygen.Thisprocessisnowrecalledas the‘‘Warburgeffect’’. Glycolysistakespartin thecytoplasm. It leadsto the NADH and ATP production through conversion of glucoseintopyruvate.Oxidativephosphorylation generatesATP intomitochondria.InrestingconditionsOXPHOSispredominant andmoreefficient.Evenif,cells withhighratesofproliferation tendtoswitchtoglycolysis.Cancercellsmakeuseofthepentose phosphatepathway(PPP),whichoriginatesfromabypassinthe glycolysis[4].PPPisnecessaryforlipidandnucleicacidsynthesis and represents an important source of NADPH.NADPH is also necessary for the production of glutathione (GSH), the major intracellularantioxidant.Thus,thePPPleadstoproliferatefastand toprotectionagainstoxidativestress[5].
P53isthemostimportantobstaclefortumorprogression.The p53homologueispresentinunicellularorganisms.Thisindicates that the tumor suppression may not be p53’s original function
[6]. BesidesDNAdamageand oncogeneactivation, nutrientflow changesactivatep53[7].Recentstudiesshowthatp53controlsthe metabolicswitchbetweenglycolysisandoxidativephosphorylation.
1.2. P53andglucosemetabolism
P53inhibitsglycolysisbyactingatdifferentlevels.Ithasbeen demonstratedthatp53decreasesglucoseimportbyreducingthe expressionof GLUT1 and GLUT4 glucose transporters [8]. P53, throughtheinhibitionofIKK,alsoregulatestheGLUT3expression
[9]. Bensaad and co-workers demonstrated that p53 reduces glycolysis by promoting TIGAR expression [5]. TIGAR depho-sphorylates fructose-2,6-bisphosphate to fructose-6-phosphate andblocksthebreakdownofglucoseintopyruvate.Thisinturn promotestheswitchtotheoxidativepentosephosphatepathway (ox-PPP).The latterleads toNADPHproduction and toa more effective protection against oxidative stress. P53-independent TIGAR accumulation is a hallmark of several tumors. Yet, the scenarioiscomplexsince p53inhibitstheglucose-6-phosphate dehydrogenase(G6PDH)expression.G6PDHtakespartinthefirst andlimitingstepoftheox-PPP[4].Themechanismbywhichp53 inhibits G6PDH represents a way to block ox-PPP in case of accumulationofTIGARincancercells.TAp73,amemberofthep53 family,inducestheexpressionofG6PDHgenesandleadstocell proliferation[10]. In mouse embryonicfibroblasts, p53inhibits glycolysis.In these cells, P53 reduces theprotein levels of the glycolyticenzymephosphoglyceratemutase(PGM).PGMconverts 3-phosphoglycerateinto2-phosphoglycerate[11].However,p53 promotesglycolysisinmusclebyinducingtheexpressionofPGM Misoform[12]andofhexokinaseII(HK2)[13].Thelatterconverts glucoseintoglucose-6-phosphate in thefirststep ofglycolysis. Thisindicatesthattheeffectsofp53onglycolysisareremarkable andtissue-specific.
1.3. P53modulatestheglucosemetabolismwhilecontrollingthe gluconeogenesis
Gluconeogenesisproduces glucoseandisessentialfortumor cellgrowth.P53repressesthegluconeogenesisbypromotingthe
expression of histone deacetilase sirtuin 6 (SIRT6). SIRT6 deacetylates forkhead box protein O1 (FOXO1). In turn, this repressestheexpressionofglucose-6-phosphatase(G6PC)andof phosphoenolpyruvate carboxykinase (PCK1). Both enzymes are rate-limitingproteinsforgluconeogenesis[14].
1.4. P53andoxidativephosphorylation
Whilerepressingglycolysis,p53promotesoxidative phosphor-ylation at distinct levels. In presence of oxygen, the pyruvate derivedfromglycolysisisconvertedinAcetylcoenzymeA (Acetyl-CoA).Thelattertakespartinthetricarboxylicacid(TCA)cycle.In TCAcycle,ATP isgenerated throughoxidativephosphorylation. Besides,TCAcycleprovidesprecursorsforanabolicpathways,so supporting cell growth and proliferation. P53-deficient cells produceless ATPfromtheTCAcyclecompared tocontrolcells
[15].Indetail,p53promotesTCAcyclebyreducingthepyruvate dehydrogenasekinase2(PDK2)expression.PDK2inactivatesthe pyruvatedehydrogenasecomplex(PCD).Inturnthispromotesthe conversionofpyruvateintolactateinsteadofAcetyl-CoA[16].
P53enhancesgenetranscriptionofmitochondrialcomponents, including subunit 1 of cytochrome c oxidase (COI) [17], and cytochromecoxidase2(SCO2).SCO2inturnregulatesthesubunit 1ofcomplexIV[15].Besides,p53inducestheexpressionofthe mitochondrial apoptosis-inducing factor (AIF) [18]. AIF acts as NADH/NADPH oxidase. AIF is also essential for the proper functioningofcomplexI.
P53reducestheexpressionoftheTCAcycle-associatedmalic enzymesME1andME2.TheseenzymesareimportantforNADPH production, lipogenesis and glutamine metabolism. Thus, P53 regulatesboththeprogressionofbiosyntheticpathwaysandthe antioxidant response [19]. Besides, p53 enhances OXPHOS by promotingthetranscriptionofglutaminase2(GLS2)[20,21].GLS2 stimulatestheproductionofglutamateand
a
-ketoglutarate.The latter is a key component of the TCA cycle involved in ATP productionandtheantioxidantresponse.1.5. P53lipidmetabolismandmitochondrialhomeostasis
Besidesalterationsinglycolysisandoxidativephosphorylation, cancer cellsshowa deregulatedlipidmetabolism.In particular, cancer cellssynthesizefattyacids.Thefattyacidsare themain reserveoflipids.Lipidsarenecessaryformembraneformationand signalingtransduction[22].Breast,colonandprostatecancercells
[23–25] have high levels of fatty acids synthases (FASN). This observation supportstherole ofFASN in tumorigenesis.In line with this, FASN inhibition counteracts cellular transformation (reviewedin[26]).FASNdeficiencycounteractscancergrowthin cellswithactivatedPI3Ksignaling.Yet,blockingFASNin K-Ras-drivencancercellshasnoeffectonproliferation[27].
P53 attenuates the fatty acids synthesis, repressing the expression of the transcriptional regulator SREBP-1 (sterol regulatoryelement-bindingprotein1c).Thelatterpromotesthe expression of triglyceride synthesis and lipogenic enzymes
[28].Thus,p53inhibitstheexpressionofFASNand(ATPcitrate lyase)ACLYgenes.Thereby,p53mayrepresstumorproliferation whileinhibitingthefattyacidssynthesis.Thefattyacidoxidation leadstotheformationofAcetyl-CoA,whichtakespartintheTCA cycle,producing ATP,NADPHandFADPH.Asmentioned above, TCA cycle is active in resting conditions. While, NADPH is necessaryforthecellularprotectionagainstoxidativestress.Upon glucosedeprivation,p53inducestheexpressionofLpin1.Thisin turnstimulatesthefattyacidsoxidation[29].Lpin1isanuclear transcriptional co-activator. Following glucose starvation Lpin1 inducestheexpressionofgenesinvolvedinfattyacidsoxidation. But,Lpin1inhibitsthefattyacidoxidationathighglucose.
S.Gonflonietal./BiochemicalPharmacologyxxx(2014)xxx–xxx 2
GModel
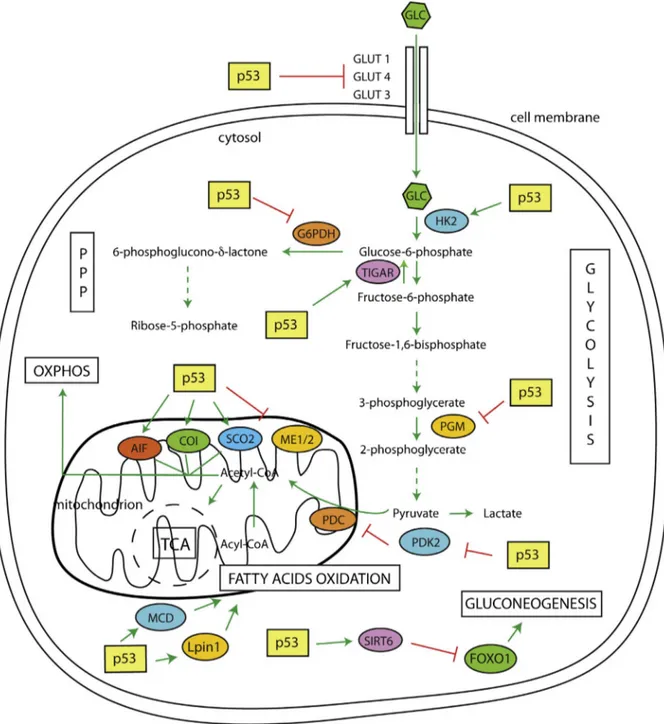
In mice, P53 controls the hepatic fatty acid oxidation in responsetofasting[30].P53inducesthetranscriptionof malonyl-CoA decarboxylase (MCD). MCD stimulates the malonyl-CoA turnover and increases carnitine palmitoyl transferase activity. Thisleadstomitochondrialfattyaciduptake(Fig.1).
In brief, P53 enhances the oxidative respiration in cells by promoting the fatty acid oxidation. In this manner, p53 may counteracttheWarburgeffect.
Mitochondrial DNA (mtDNA) contains genes of enzymatic complexesinvolvedinOXPHOS.P53protectsmitochondrialDNA frommutations,promotingthetranscriptionoftheribonucleotide reductase p53R2. Besides, p53 preserves mtDNA integrity and regulatesthemtDNAcopynumber[31].Thereby,p53promotes
theoxidativephosphorylationwhileensuringmitochondrialDNA protection.
Yet,itremainsanopenquestionwhether(andtowhatextent) the metabolic status may contribute to genomic maintenance. Recentevidencesuggeststhatgenomicstabilityrequires coopera-tionofp53andSIRT1.
2. EssentialeffectsofSIRT1onmetabolismandgenome protection
SirtuinsareafamilyofclassIIIdeacetylases(NAD+-dependent).
These enzymes control many cellular pathways, including metabolism,stressandgenomestability.
Fig.1.p53inhibitsglycolysisbyregulatingseveralcomponentsofthisprocess.P53inhibitsglucosetransporter(GLUT1-4-3)andtheglycolyticenzymePGM.Moreover,p53 inhibitsglycolysisbypromotingTIGARexpression.Atthesametime,p53inhibitstheox-PPPbyactingonG6PDH.Thep53roleistissue-specific.Inmusclep53enhances glycolysisbypromotingthetranscriptionofHK2.ThroughtheexpressionofSIRT6,p53reducesthegluconeogenesis.p53stimulatestheoxidativephosphorylationby inducingtheexpressionofseveralimportantcomponentsofthismetabolicprocess(i.e.AIF,COIandSCO2andmalicenzymesME1/2).Moreover,p53promotestheOXPHOS byreducingtheexpressionofPDK2.Inturn,thispromotestheproductionofAcetyl-CoAinsteadoflactate.Lastly,inresponseofnutrientstarvation,p53promotesthe expressionofLpin1andMCD,stimulatingtheconversionofAcyl-CoAtoAcetyl-CoA.Inthiswayp53stimulatestheoxidativephosphorylationinsteadofglycolysis.
ThebasicfunctionofSirtuinslinkschromatindynamics/gene expressiontoenvironmentalstimuli. Atthesame time,Sirtuins ensure genome protection [32]. Genome integrity relies on chromatin structure and DNA repair mechanisms. Chromatin andDNArepairdependbydeacetylationofhistones(andofother chromatin-associatedfactors)[33,34].Sirtuinswerediscoveredin yeast (Silent Information Regulator, Sir2). They were genes necessaryfortherepressionofsilent-matingtypeloci [35].The mammalianfamilyofSirtuinscomprisesofsevenproteins, SIRT1-SIRT7. These enzymesare ubiquitously expressed. SIRT1 is the best-studiedgene and thelargestin sequence. NAD+, allosteric
modulators,nucleo-cytoplasmshuttling,transcriptionregulators activateSIRT1[36].ThemammalianSirtuinsarepresentindistinct subcellularcompartments.Some Sirtuinsarepresentinto mito-chondria.Thisimpliesa role ofSirtuinsformetabolismand for mitochondriahomeostasis[37].
2.1. Metabolismandsignaling
Metabolism and signaling pathways are interconnected
[38].Cancercells exploitthesignalling-dependentregulationof metabolismtofuel macromolecular biosynthesis. Thissupports their fast growth [39–41]. Nutrient-sensing molecules are the AMP-activated kinases (AMPK) and the mammalian target of rapamycin complex 1 (mTORC1). These enzymes are key regulatorsofthemetabolicstatusofthecells.Besides,metabolites generateposttranslationalmodifications(PTMs)[42].
CytosolicandnuclearAcetyl-CoAlevelsmodulatePTMs,gene expression,signalingcascadesandmetabolism[38].Twoopposite enzymesasacetyltransferasesanddeacetylasesregulate acetyla-tion.Thelatteris a PTMinducedbythemetabolite Acetyl-CoA
[43,44].Intracellularmetabolicconditionscontroltheactivityof thesetwoclassesofenzymes.SIRT1deacetylasedependsonNAD+
as a cofactor. Besides NAD+, NAD+ biosynthetic enzymes (as
Nicotinamidephosphoribosyltransferase–Nampt)promoteSIRT1 deacetylaseactivity.
Proteomic studiesindicatedthatacetylation(andAcetyl-CoA production)regulatesseveralsignalingevents.Emergingconcepts
suggestthatacetylationislikea‘‘sensor’’ofAcetyl-CoAavailability withincells.ATP-citratelyase(ACL)andAcetyl-CoAsynthetase1 (ACECS1) are essential enzymes for Acetyl-CoA production. ACECS1isdeacetylated(andactivated)bySIRT1under nutrient-limited conditions [45]. Acetylation of ACECS1 may serve as a regulatory/inhibitory modification in response to metabolic changes. Many enzymes involved in metabolism are also acetylated [46,47]. Anacetyl-mimic mutant of pyruvate kinase (PKM2)promotestumorxenografsgrowth[48].
Besides,Acetyl-CoAproductionenhances histone acetylation and so gene transcription [49]. Nutrient availability, metabolic conditions and posttranslational modifications of the enzymes controlAcetyl-CoAproduction.PhosphorylationofACLbyAKTand by PKA increases ACL activity [50–52]. Cancer cells have the phosphoinositide3-kinase(PI3K)-AKTpathwayactivated. Acetyl-CoA production is persistent, providing a continuous signal to promotegrowth[38].PTMsofdeacetylasesarelinkedto Acetyl-CoAproduction.SIRT1modificationsexerteitherstimulatoryor inhibitory effects on SIRT1 activity [53]. Different kinases/ modifiers anddiverse multi-siteregulatorymechanismscontrol SIRT1 activity [54]. In response to oxidative stress, c-Jun N-terminal kinase (JNK) phosphorylates SIRT1 at three sites
[55]. These modifications increase SIRT1 activity. In contrast, mTORphosphorylatesSIRT1atasinglesite(S47)inresponseto oxidativestress.ThismodificationinhibitsSIRT1activity.AMPK targets humanSIRT1 at T344.This modification reduces SIRT1 capabilitytodeacetylatep53[56,57].OtherkinasesasCK2,DYRK1 andDYRK3phosphorylateSIRT1indifferentsites.These modifica-tionsinturnstimulateSIRT1activity[58–60].Incontrast,CyclinB/ Cdk1(acellcycledependentkinase)phosphorylatesSIRT1atT530 andS540.BothmodificationsinhibitSIRT1activity[61].
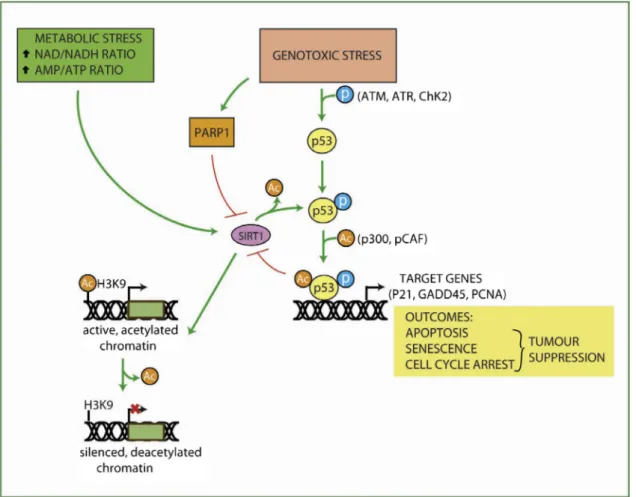
Thefirstdiscoverednon-histonesubstrateofSIRT1isp53.P53 playsacentralroleinSIRT1-mediatedsenescenceandintumor progression[62].SIRT1repressesp53-dependenttransactivation intumorsorinmouseembryonicfibroblasts[63].Yet,p53binding toSIRT1promoterrepressesSIRT1 transcription[62].Thus, p53 providesafeedbackcircuiteithertoregulateSIRT1expressionand thep53response. Lownutrientconditions increaseNAD/NADH ratio, promoting SIRT1 activity. PARP1 activity leads to NAD+ depletion, limitingSIRT1 activity[64].Moreover, SIRT1 induces gene silencing through H3K9 deacetylation of target gene promoters(Fig.2).
SIRT1reducesthep53-dependentapoptosisinducedbystress
[65,66].SIRT1-deficientmiceshowp53hyperacetylationfollowing DNA damage. They also showan increasedionizing radiation-induced apoptosis in thymocytes [66]. Other studies report a modest effect of SIRT1 on the p53-mediated responses
[67,68]. These opposite results imply the existence of other mechanismsbywhichSIRT1exertsitseffectonp53.Convincing evidence indicates that SIRT1 regulates the p53 subcellular localization.Likely,SIRT1preventsp53nucleartranslocation.This inturnpromotescytosolicaccumulationofp53anditspassageto mitochondria.Inbrief,SIRT1blocksp53transcription-dependent apoptosis. At the same time SIRT1 increases p53-mediated (transcription-independent)apoptosis[62].
SIRT1 was first considered as a potential tumor promoter, repressing p53. Yet, recent results indicated that Sirt1 acts as tumorsuppressorbyfacilitatingmitochondria-dependent apopto-ticresponse.
SIRT1deacetylatesdifferenthistones(H4,H3,andH1)andalso manynon-histoneproteins.SIRT1substratestakepartindiverse cellularresponses[69,70].SIRT1 substratescan be1) histone-modifyingenzymessuchasSUV39H1,PCAF,p300andTIP60;2) transcription factors regulating cell cycle progression/survival under various stress conditions (p53, nuclear factor (NF)-
k
B, p73, forkhead transcription factors FOXO1, FOXO3a, Myc,Table 1 Sirtsubstrates.
Substrates Cellularfunction References
H4 Histones PMID:17694090
H3 PMID:19411844
H1 PMID:15469825
SUV39H1 Histone-modifyingenzymes PMID:21504832
PCAF PMID:19188449
p300 PMID:15632193
TIP60 PMID:22586264
p53 Transcriptionfactors PMID:11672523
E2F1 PMID:16892051 NFkB PMID:15152190 P73 PMID:16998810 FOXO1 PMID:25009184 FOXO3a PMID:14976264 Myc PMID:21807113
HIF-1a/HIF2-a PMID:24423936
ATM Cellsignalingcomponents PMID:23852118
AMPK PMID:19276888
AKT PMID:24436432
Ku70 DNArepairmodulators PMID:23247197
WRN PMID:18203716
Rad51 PMID:20097625
NBS1 PMID:17612497
APE1 PMID:19934257
BRCA1 PMID:18851829
PGC-1a Regulatorsofmetabolism PMID:15744310
CLOCK PMID:24442997
PER2 PMID:18662546
PPARg PMID:15175761
S.Gonflonietal./BiochemicalPharmacologyxxx(2014)xxx–xxx 4
GModel
hypoxia-inducibletranscriptionfactors(HIF)-1
a
andHIF2-a
);3) cell signaling components (modifiers, enzymes) AKT; 4) DNA repairmodulatorssuchasKu70,WRN,NBS1,APE1;5)regulators ofmetabolism,circadian clockincludingPGC-1a
,PER2 [71,72](seeTable1).
Mousemodelssupportthemaintenanceofgenomicstabilityas the fundamental role of SIRT1 [32]. Yet, SIRT1 modulates the regulatorysignalingcircuitsofcellhomeostasis.SIRT1maydeal withoxidativestress,aging,metabolism,andgenomeprotection. Todate,theidentificationofSirtuin-regulatedsignalingtargets remainsincomplete.Reversaloftheacetylationaffects transcrip-tionfactors [66]and their nuclear localization.Yet, removalof acetylationmayinduceproteinbreakdown[73].
Acetylationofthemitogen-activatedproteinkinasekinase-1 (MEK1) enhances its kinase activity. Treatment with Sirtuin inhibitorsorsiRNAsilencingofSIRT1orSIRT2,enhancesMEK1 acetylation.Thismodificationcausestheconsequent phosphor-ylationofextracellularsignal-regulatedkinase(ERK).An acetyl-mimicmutantofMEK1promotesinappropriategrowth proper-ties. This indicates that acetylation of MEK1 may exert an oncogenicpotential[74].ItisofinteresttopointoutthatSIRT2is anewAKTadaptorrequiredforoptimalAKTactivity. Constitu-tiveSIRT2bindingtoAKT isdependentonphosphorylationat T101 by AMP-activated kinase. Pharmacological inhibition or genetic down-regulation of SIRT2 inhibits AKT activation in stimulatedcells.Ofnote,overexpressionofSIRT2enhances AKT-mediated pathway activation. In cells with constitutive PI3K activation, AKT interacts with a nuclear Sirtuin, (SIRT1). But,
inhibition of PI3K induces SIRT1 dissociation and increased associationwithSIRT2.ThissupportstheuseofSIRT1andSIRT2 inhibitorsinthetreatmentoftumorcellswithanincreasedPI3K activity[75].
Reversibleacetylationcontrols thesubcellularlocalization of manyothersignalingfactors/components.Besides,itregulatesthe silencingofgeneexpression.SIRT1promotesnuclearretentionof High-mobilitygroup(HMG)B1.SIRT1modulatesdamagesignaling initiatedbyHMGB1secretionuponstress[76].SIRT1reducesthe expressionofsurvivin,throughH3K9deacetylationonitspromoter
[77].
SIRT1 may promote and/or suppress tumorigenesis
[32,78,79]. On one hand, SIRT1-deficiency may help tumor progression,promotinggenomicinstability.Yet,tumorcellstend toneedofSirtuinstoproliferate,torepairwithlowfidelityandto evolve[78].ThebifurcatedroleofSIRT1intumorigenesisisdueto itspeculiarroleingenomeprotection[78].
2.2. Genomicstability
SIRT1actsonthegenomethroughvariousmechanismsandat differentlevels.SIRT1regulateschromatinmodifications, facilitat-ing DNA repair. Besides, SIRT1 connects energy and metabolic flowstochromatindynamicsandgeneexpression.
SIRT1 facilitatesboth constitutiveheterochromatin(CH) and facultative heterochromatin (FH) formation. SIRT1 enhances methyl-transferases activity. This promotesthe methylation of CpGislands(reviewedin[79]).
Fig.2.Followinggenotoxicstresses,p53isphosphorylatedatS15byATM,ATR,Chk2kinases.Besides,p53ismodifiedatK382byp300andPCAFacetyltransferases.These posttranslationalmodificationsstimulatep53transcriptionofspecifictargetgenes(p21,GADD45,PCNA).Inturnthispromotescellcyclearrest,apoptosisorsenescence.P53 bindingtoSIRT1promoterrepressesSIRT1transcription.Bythismechanism,p53providesafeedbackcircuittoregulateSIRT1expressionandatthesametime,tomodulate p53-transcriptiondependentresponse.LownutrientconditionsactivateSIRT1.While,PARP1activityleadstoNAD+depletion,limitingSIRT1activity[64]. Moreover,SIRT1 inducesgenesilencingthroughH3K9deacetylationoftargetgenepromoters.
SIRT1inducesepigeneticsilencingoftargetgenes[80,81].SIRT1 modifies histone (H4K16Ac, H3K9Ac, H1K26Ac) and chromatin factors(asthehistonemethyl-transferaseSUV39H1).LossofSIRT1 prevents both the spreading of heterochromatin marks (H3K79me3)andlocalizationofheterochromatinprotein1(HP1)
[82].TheinterplaybetweenSIRT1andSUV39H1ismediatedbythe E3ubiquitinligaseMDM2.SIRT1inhibitsSUV39H1 polyubiquitina-tion by MDM2 and so increases SUV39H1[83]. High levels of SUV39H1invivoenhanceitsturnoverinpericentromeric hetero-chromatin regions. This ensures genome integrity. SIRT1 may generate aberrant methylation of the CpG islands in promoters following DNA damage. This promotes heritable gene silencing
[84]. SIRT1-mediated gene silencing may affect both tumor promotingandtumorsuppressorgenes.SIRT1inhibitionreactivates silencedtumorsuppressorgenesincancercell.Thisoccurswithout lossofpromoterhypermethylation[85].
2.3. DNArepair
SIRT1takespartinDNAsignalingandrepairatdifferentlevels. For the repairof singlestrand breaks (SSBs), SIRT1 controls the nucleotideexcisionrepair(NER)pathway.SIRT1interactswithXPA (xerodermapigmentosumcomplementationgroupA)[86].SIRT1 enhancesXPC(xerodermapigmentosumcomplementationgroup C)expression.SIRT1preventsAKT-mediatednuclearlocalizationof XPC transcriptional repressors. Inhibition of SIRT1 affects XPC transcription, impairing NER function [87]. Several DNA repair factorsarealsosubstratesofSIRT1,amongthemX-rayrepair cross-complementinggene4protein(XRCC4p)[88];Ku70[89];Werner’s Syndrome protein (WRN)[90]. SIRT1 binds to and modifies Nijmegenbreakagesyndromeprotein(NBS1).NBS1isaregulatory component of the MRE11-Rad51-NBS (MRN) nuclear complex
[91,92].MRNcomplexworks asa‘‘sensor’’inthe earlystagesof doublestrandbreak(DSB)signaling.MRNpromotestheformation of
g
-H2AXfociupong
-irradiation[93].SIRT1recruitmenttoDSBs dependsonATM-mediatedsignaling(i.e.g
-H2AXphosphorylation)[94].SIRT1interactswith(andmodifies)twomembersoftheMYST acetyltransferase family (hMOF and TIP60). Both enzymes are necessaryforcellgrowth,DNArepairandapoptosis[95–97].SIRT1 inhibits their activity by promoting their ubiquitin-mediated degradation.FollowingDNAbreaks,SIRT1bindingisreducedand hMOFandTIP60takepartinthedamageresponse[97–99].SIRT1is alsoconnectedtothenon-homologousendjoining(NHEJ)repair pathway,throughtheproteinKu70.DeacetylationofKu70bySIRT1 preventsKu70 translocation.At the sametime, SIRT1promotes Ku70-dependentDNArepair[100–102].
3. SIRT1-p53:nodesconnectingmetabolismandgenome integrity
Mousemodelssupporta roleofSIRT1forgenomeprotection
[66,93,103,104].SIRT1regulatesp53acetylation.SIRT1-deficient thymocytesexhibitp53hyperacetylation[66].Doubleknockout miceshowthatgenomicintegrityneedscooperationbetweenp53 andSIRT1[93].
In mice,evidenceshowsthatp53tumorsuppressorfunction reliesonp53abilitytomodulatemetabolism[105].Besides,the p53 family members p63 and p73 are emerging as critical components in the regulation of metabolism [106,107]. This implies that the metabolic functions of p63/p73 may also contributetotumorsuppression.Yet,theseeffectsremainelusive inparticularwithtumorsexpressingmutantformsofp53[108]. SIRT1 modulatesbothp53transcription-dependentand p53-independentapoptosis.Thelatterisinitiatedthroughcytochrome creleasefromthemitochondria.Emergingevidenceindicatesthat SIRT1preventsp53nucleartranslocation.SIRT1redirectsp53from
the cytosol tothe mitochondria in response to increased ROS. Likely,thebiologicaleffectofp53enrichmentintomitochondria inducestranscription-independentapoptosis.Inthisway, oxida-tive stress modulates cell fate through SIRT1-mediated p53 deacetylation(reviewedin[62]).
DNA Damage stress Response (DDR) relies on cell cycle checkpoints for repairing DNA lesions. Failure to balancing of DDR response promotes ageing and cancer. How are cellular metabolismandDDRinterconnected?Besides,therolesofp53and SIRT1remainstillundefined.
Thechemotherapeutic drugcisplatin,causesoxidative stress and various types of DNA alterations. Stechowand co-workers perform metabolic profiling by mass spectrometry (MS) of embryonicstem(ES)cells.EScellsaretreatedfordifferenttime periods with cisplatin. MS and transcriptomics analyses of cisplatin-treatedES-cellsshowthatcisplatinaltersthemetabolic pathways.Yet,severalmetabolicenzymesinducedbycisplatinare alsop53targetgenes.Theseresultsshowthatmetabolicpathways areinterconnectedwithDNAdamageresponse.Itisofinterestthat p53playsacentralroleforbothprocesses[109].
Emerging evidenceindicates that metabolic changes have a direct effect on chromatin and DNA repair. Several chromatin-modifying enzymes, involved in DNA repair, are regulated by metabolic cofactors. Thus, alterations of energy and metabolic flowshaveprofoundconsequencesongenomeintegrity.
ChromatinplaysanessentialroleinDNArepairpathways (non-homologous end-joining (NHEJ) or homologous recombination (HR)). Many histone modifications are necessary to assembly signaling-platformsforrepairmechanisms.Histonemodifications (includingphosphorylation,acetylation,methylation, ubiquityla-tion)regulateDSBrepair[110–114].Enzymes,involvedinhistone modifications,needmetabolitesascofactorsorsubstratesfortheir activity. Metabolic cofactors/substrates including Acetyl-CoA (involved in Acetyl-CoA-dependent histone acetylation) and NAD+ modulate histone modifications. NAD+ is essential for
NAD+-dependent deacetylation or NAD+-dependent
polyADP-ribosylation of chromatin. Besides, S-adenosyl-L-methionine
(SAM)andFAD/
a
-ketoglutarateinducemethylationand demeth-ylation of histones and/or DNA. Metabolic modifications may contributetoDNAdouble-strand(DSBs)repairdefects[115].At whatextentdothemetabolicchangesinterferewith chromatin-directed repair processes? It remains an open question. These studiesrepresentanexcitingareaofinvestigation.4. Conclusionsandperspectives
Cancer is a complex disease driven by epigenetic changes, geneticmodificationsinoncogenes andtumorsuppressors.Yet, alterations in metabolism have profound effects on the cell growth/survival.Cellhomeostasisrequiresfine-tuningofenergy and metabolic flows withgenomestability. P53 and SIRT1 are nodesregulatingmetabolism,DNArepairandsenescence.
Metabolic p53 responsesare important for maintaining cell homeostasis and for preventing tumor development. Metabolic andotherstresssignalsactivatep53[116].Recently,otherauthors reviewed the cytosolic functions of p53 [117]. P53 regulates metabolismandinducessenescence.Thisrepresentsaparadigm fortargetingofmetabolicalterationsassociatedwithcancer[118]. RecentevidenceindicatesadualeffectofSIRT1oncancer.SIRT1 enhancescellsurvival,allowingindefinitecelldivision.Atthesame time,SIRT1promotesgenomestabilityunderstressconditions[79]. DNArepairneedsdynamicchromatinremodeling,re-assembly of nucleosome, histone variant exchanges, signaling response. Metabolite-associatedchangesatchromatinmaypromote malig-nant transformation through deregulation of DNA repair
[115].Howdometabolicchangesinterferewithchromatin-directed
S.Gonflonietal./BiochemicalPharmacologyxxx(2014)xxx–xxx 6
GModel
repairprocesses?Itremainselusive.Thein-depthunderstandingof metabolite effects on DNA repair will help the developmentof combinedtargetedtherapiestoeradicatecancer.
Acknowledgements
We acknowledge support from AIRC (Italian Association for Cancer Research, IGgrant 2011No. 11344)to SG. Research in M.D.’slab is supported bythe‘‘Recherche Canceret Sang’’,the ‘‘RecherchesScientifiquesLuxembourgassociation,the‘‘EenHaerz firkriibskrankKanner’’association,theActionLions‘‘Vaincrele Cancer’’associationandbyTe´le´vieLuxembourg.MDissupported bytheNationalResearchFoundationofKorea(NRF)grantforthe Global Core Research Center (GCRC) funded by the Korea Government,Ministryof Science,ICT & FuturePlanning(MSIP) (No.2011-0030001).
References
[1]VousdenKH,PrivesC.Blindedbythelight:thegrowingcomplexityofp53. Cell2009;137:413–31.
[2]WangDB, KinoshitaC,KinoshitaY,MorrisonRS.p53 andmitochondrial functioninneurons.BiochimicaetBiophysicaActa2014;1842(8):1186–97. http://dx.doi.org/10.1016/j.bbadis.2013.12.015.
[3]WarburgO,WindF,NegeleinE.Themetabolismoftumorsinthebody.The JournalofGeneralPhysiology1927;8:519–30.
[4]JiangP,DuW,WangX,MancusoA,GaoX,WuM,etal.p53regulates biosynthesisthroughdirectinactivationofglucose-6-phosphate dehydroge-nase.NatureCellBiology2011;13:310–6.
[5]BensaadK,TsurutaA,SelakMA,VidalMN,NakanoK,BartronsR,etal.TIGAR, ap53-inducibleregulatorofglycolysisandapoptosis.Cell2006;126:107–20. [6]LuWJ,AmatrudaJF.AbramsJM.p53ancestry:gazingthroughan
evolution-arylens.NatureReviews.Cancer2009;9:758–62.
[7]MaddocksOD,VousdenKH.Metabolicregulationbyp53.Journalof Molecu-larMedicine(BerlinGermany)2011;89:237–45.
[8]Schwartzenberg-Bar-YosephF,ArmoniM,KarnieliE.Thetumorsuppressor p53down-regulatesglucosetransportersGLUT1andGLUT4geneexpression. CancerResearch2004;64:2627–33.
[9]KawauchiK,ArakiK,TobiumeK,TanakaN.p53regulatesglucosemetabolism throughanIKK-NF-kappaBpathwayandinhibitscelltransformation.Nature CellBiology2008;10:611–8.
[10]DuW,JiangP,MancusoA,StonestromA,BrewerMD,MinnAJ,etal.TAp73 enhancesthepentosephosphatepathwayandsupportscellproliferation. NatureCellBiology2013;15:991–1000.
[11]KondohH,LleonartME,GilJ,WangJ,DeganP,PetersG,etal.Glycolytic enzymescanmodulatecellularlifespan.CancerResearch2005;65:177–85. [12]Ruiz-LozanoP,HixonML,WagnerMW,FloresAI,IkawaS,BaldwinJrAS,etal. p53isatranscriptionalactivatorofthemuscle-specificphosphoglycerate mutasegeneandcontributesinvivotothecontrolofitscardiacexpression. CellGrowth&Differentiation:TheMolecularBiologyJournaloftheAmerican AssociationforCancerResearch1999;10:295–306.
[13]MathupalaSP,KoYH,PedersenPL.HexokinaseII:cancer’sdouble-edged swordactingasbothfacilitatorandgatekeeperofmalignancywhenboundto mitochondria.Oncogene2006;25:4777–86.
[14]ZhangP,TuB,WangH,CaoZ,TangM,ZhangC,etal.Tumorsuppressorp53 cooperateswithSIRT6to regulategluconeogenesisby promotingFoxO1 nuclearexclusion.ProceedingsoftheNationalAcademyofSciencesofthe UnitedStatesofAmerica2014;111:10684–89.
[15]MatobaS,KangJG,PatinoWD,WraggA,BoehmM,GavrilovaO,etal.p53 regulatesmitochondrialrespiration.Science2006;312:1650–3.
[16]ContractorT,HarrisCR.p53negativelyregulatestranscriptionofthe pyru-vatedehydrogenasekinasePdk2.CancerResearch2012;72:560–7. [17]OkamuraS,NgCC,KoyamaK,TakeiY,ArakawaH,MondenM,etal.
Identifi-cationofsevengenesregulatedbywild-typep53inacoloncancercellline carryingawell-controlledwild-typep53expressionsystem.Oncology Re-search1999;11:281–5.
[18]VahsenN,CandeC,BriereJJ,BenitP,JozaN,LarochetteN,etal.AIFdeficiency compromisesoxidativephosphorylation.TheEMBOJournal2004;23:4679–89. [19]JiangP,DuW,MancusoA,WellenKE,YangX.Reciprocalregulationofp53 and malic enzymes modulates metabolism and senescence. Nature 2013;493:689–93.
[20]HuW,ZhangC,WuR,SunY,LevineA,FengZ.Glutaminase2,anovelp53 targetgeneregulatingenergymetabolismandantioxidantfunction. Proceed-ingsoftheNationalAcademyofSciencesoftheUnitedStatesofAmerica 2010;107:7455–60.
[21]SuzukiS,TanakaT,PoyurovskyMV,NaganoH,MayamaT,OhkuboS,etal. Phosphate-activatedglutaminase(GLS2),ap53-inducibleregulatorof gluta-minemetabolismandreactiveoxygenspecies.ProceedingsoftheNational AcademyofSciencesoftheUnitedStatesofAmerica2010;107:7461–6.
[22]SantosCR,SchulzeA.Lipidmetabolismincancer.TheFEBSJournal2012;279: 2610–2623.
[23]LiJN,MahmoudMA,HanWF,RippleM,PizerES.Sterolregulatory element-bindingprotein-1participatesintheregulationoffattyacidsynthase expres-sionincolorectalneoplasia.ExperimentalCellResearch2000;261:159–65. [24]SwinnenJV,VanderhoydoncF,ElgamalAA,EelenM,VercaerenI,JoniauS,
etal.Selectiveactivationofthefattyacidsynthesispathwayinhuman prostatecancer.InternationalJournalofCancer.Journalinternationaldu cancer2000;88:176–9.
[25]YoonS,LeeMY,ParkSW,MoonJS,KohYK,AhnYH,etal.Up-regulationof acetyl-CoAcarboxylasealphaandfattyacidsynthasebyhumanepidermal growthfactorreceptor2atthetranslationallevelinbreastcancercells.The JournalofBiologicalChemistry2007;282:26122–31.
[26]MenendezJA,LupuR.Fattyacidsynthaseandthelipogenicphenotypein cancerpathogenesis.NatureReviews.Cancer2007;7:763–77.
[27]YellenP,FosterDA.Inhibitionoffattyacidsynthaseinducespro-survivalAkt andERKsignalinginK-Ras-drivencancercells.CancerLetters2014;353(2): 258–263.http://dx.doi.org/10.1016/j.canlet.2014.07.027.
[28]YahagiN,ShimanoH,MatsuzakaT,NajimaY,SekiyaM,NakagawaY,etal. p53Activationinadipocytesofobesemice.TheJournalofBiological Chem-istry2003;278:25395–400.
[29]AssailyW,RubingerDA,WheatonK,LinY, MaW,XuanW,etal. ROS-mediatedp53inductionofLpin1regulatesfattyacidoxidationinresponseto nutritionalstress.MolecularCell2011;44:491–501.
[30]LiuY,HeY,JinA,TikunovAP,ZhouL,TolliniLA,etal.Ribosomal protein-Mdm2-p53pathwaycoordinatesnutrientstresswithlipidmetabolismby regulatingMCDandpromotingfattyacidoxidation.ProceedingsoftheNational AcademyofSciencesoftheUnitedStatesofAmerica2014;111:E2414–22. [31]KulawiecM,AyyasamyV,SinghKK.p53regulatesmtDNAcopynumberand
mitocheckpointpathway.JournalofCarcinogenesis2009;8:8.
[32]Bosch-PresegueL,VaqueroA.Sirtuinsinstressresponse:guardiansofthe genome. Oncogene 2014;33(29):3764–75. http://dx.doi.org/10.1038/onc. 2013.344.
[33]VaqueroA,SternglanzR,ReinbergD.NAD+-dependentdeacetylationofH4 lysine16byclassIIIHDACs.Oncogene2007;26:5505–20.
[34]Vaquero A.Theconserved roleofsirtuinsinchromatinregulation. The InternationalJournalofDevelopmentalBiology2009;53:303–22. [35]BlanderG,GuarenteL.TheSir2familyofproteindeacetylases.AnnualReview
ofBiochemistry2004;73:417–35.
[36]SatohA,SteinL,ImaiS.Theroleofmammaliansirtuinsintheregulationof metabolism,aging,andlongevity.HandbookofExperimentalPharmacology 2011;206:125–62.
[37]ZhongL, Mostoslavsky R. Fine tuningour cellularfactories: sirtuinsin mitochondrialbiology.CellMetabolism2011;13:621–6.
[38]WellenKE,ThompsonCB.Atwo-waystreet:reciprocalregulationof metab-olismandsignalling.NatureReviews.MolecularCellBiology2012;13:270–6. [39]DeBerardinisRJ, Lum JJ,Hatzivassiliou G, ThompsonCB. Thebiologyof cancer:metabolicreprogrammingfuelscellgrowthandproliferation.Cell Metabolism2008;7:11–20.
[40]KoppenolWH,BoundsPL,DangCV.OttoWarburg’scontributionstocurrent conceptsofcancermetabolism.NatureReviews.Cancer2011;11:325–37. [41]VanderHeidenMG,CantleyLC,ThompsonCB.UnderstandingtheWarburg
effect:themetabolicrequirementsofcellproliferation.Science2009;324: 1029–1033.
[42]MetalloCM,VanderHeidenMG.Metabolismstrikesback:metabolicflux regulatescellsignaling.Genes&Development2010;24:2717–22. [43]HaigisMC,SinclairDA.Mammaliansirtuins:biologicalinsightsanddisease
relevance.AnnualReviewofPathology2010;5:253–95.
[44]SchwerB,VerdinE.Conservedmetabolicregulatoryfunctionsofsirtuins.Cell Metabolism2008;7:104–12.
[45]HallowsWC,LeeS,DenuJM.Sirtuinsdeacetylateandactivatemammalian acetyl-CoAsynthetases.ProceedingsoftheNationalAcademyofSciencesof theUnitedStatesofAmerica2006;103:10230–35.
[46]KimSC,SprungR,ChenY,XuY,BallH,PeiJ,etal.Substrateandfunctional diversityoflysineacetylationrevealedbyaproteomicssurvey.Molecular Cell2006;23:607–18.
[47]ZhaoS,XuW,JiangW,YuW,LinY,ZhangT,etal.Regulationofcellular metabolismbyproteinlysineacetylation.Science2010;327:1000–4. [48]LvL,LiD,ZhaoD,LinR,ChuY,ZhangH,etal.AcetylationtargetstheM2
isoformofpyruvatekinase fordegradationthroughchaperone-mediated autophagyandpromotestumorgrowth.MolecularCell2011;42:719–30. [49]CaiL,SutterBM,LiB,TuBP.Acetyl-CoAinducescellgrowthandproliferation
bypromotingtheacetylationofhistonesatgrowthgenes.MolecularCell 2011;42:426–37.
[50]PotapovaIA,El-MaghrabiMR,DoroninSV,BenjaminWB.Phosphorylationof recombinanthumanATP:citratelyasebycAMP-dependentproteinkinase abolisheshomotropicallostericregulationoftheenzymebycitrateand increasestheenzymeactivity.AllostericactivationofATP:citratelyaseby phosphorylatedsugars.Biochemistry2000;39:1169–79.
[51]BerwickDC,HersI,HeesomKJ,MouleSK,TavareJM.Theidentificationof ATP-citratelyaseasaproteinkinaseB(Akt)substrateinprimaryadipocytes. TheJournalofBiologicalChemistry2002;277:33895–900.
[52]SaleEM,HodgkinsonCP,JonesNP,SaleGJ.Anewstrategyforstudying proteinkinaseBanditsthreeisoforms.RoleofproteinkinaseBin phos-phorylatingglycogensynthasekinase-3, tuberin,WNK1,andATPcitrate lyase.Biochemistry2006;45:213–23.
[53]Luna A,AladjemMI,KohnKW.SIRT1/PARP1crosstalk: connectingDNA damageandmetabolism.GenomeIntegrity2013;4:6.
[54]FlickF,LuscherB.Regulationofsirtuinfunctionbyposttranslational mod-ifications.FrontiersinPharmacology2012;3:29.
[55]NasrinN,KaushikVK,FortierE,WallD,PearsonKJ,deCaboR,etal.JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE 2009;4:e8414.
[56]LeeCW,WongLL,TseEY,LiuHF,LeongVY,LeeJM,etal.AMPKpromotesp53 acetylationviaphosphorylationandinactivationofSIRT1inlivercancercells. CancerResearch2012;72:4394–404.
[57]LauAW,LiuP,InuzukaH,GaoD.SIRT1phosphorylationbyAMP-activated proteinkinaseregulatesp53acetylation.AmericanJournalofCancer Re-search2014;4:245–55.
[58]KangH,JungJW,KimMK,ChungJH.CK2istheregulatorofSIRT1 substrate-bindingaffinity,deacetylaseactivityandcellularresponsetoDNA-damage. PLoSONE2009;4:e6611.
[59]ZschoernigB,MahlknechtU.Carboxy-terminalphosphorylationofSIRT1by proteinkinaseCK2.BiochemicalandBiophysicalResearchCommunications 2009;381:372–7.
[60]KangH,SuhJY,JungYS,Jung JW,KimMK,ChungJH.Peptideswitchis essentialforSirt1deacetylaseactivity.MolecularCell2011;44:203–13. [61]SasakiT,MaierB,KoclegaKD,ChruszczM,GlubaW,StukenbergPT,etal.
PhosphorylationregulatesSIRT1function.PLoSONE2008;3:e4020. [62]YiJ,LuoJ.SIRT1andp53,effectoncancer,senescenceandbeyond.Biochimica
etBiophysicaActa2010;1804:1684–9.
[63]LangleyE,PearsonM,FarettaM,BauerUM,FryeRA,MinucciS,etal.Human SIR2 deacetylatesp53and antagonizesPML/p53-inducedcellular senes-cence.TheEMBOJournal2002;21:2383–96.
[64]CantoC,AuwerxJ.InterferencebetweenPARPsandSIRT1:anovelapproach tohealthyageing.Aging2011;3:543–7.
[65]LoweSW,LinAW.Apoptosisincancer.Carcinogenesis2000;21:485–95. [66]ChengHL,MostoslavskyR,SaitoS,ManisJP,GuY,PatelP,etal.
Develop-mentaldefectsandp53hyperacetylationinSir2homolog(SIRT1)-deficient mice.ProceedingsoftheNationalAcademyofSciencesoftheUnitedStatesof America2003;100:10794–99.
[67]Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307–10.
[68]HanMK,SongEK,GuoY,OuX,MantelC,BroxmeyerHE.SIRT1regulates apoptosisandNanogexpressioninmouseembryonicstemcellsby control-lingp53subcellularlocalization.CellStemCell2008;2:241–51.
[69]BrooksCL,GuW.HowdoesSIRT1affectmetabolism,senescenceandcancer. NatureReviews.Cancer2009;9:123–8.
[70]DengCX.SIRT1,isitatumorpromoterortumorsuppressor.International JournalofBiologicalSciences2009;5:147–52.
[71]HoutkooperRH,PirinenE,AuwerxJ.Sirtuinsasregulatorsofmetabolismand healthspan.NatureReviews.MolecularCellBiology2012;13:225–38. [72]SaundersLR,VerdinE.Sirtuins:criticalregulatorsatthecrossroadsbetween
cancerandaging.Oncogene2007;26:5489–504.
[73]vanGentR,DiSanzaC,vandenBroekNJ,FleskensV,VeenstraA,StoutGJ, etal.SIRT1mediatesFOXA2breakdownbydeacetylationina nutrient-dependentmanner.PLoSONE2014;9:e98438.
[74]YeungF,RamseyCS,Popko-SciborAE,AllisonDF,GrayLG,ShinM,etal. Regulation of the mitogen-activated protein kinase kinase (MEK)-1 by NAD-dependent deacetylases. Oncogene 2014. http://dx.doi.org/10.1038/ onc.2014.39.
[75]RamakrishnanG,DavaakhuuG,KaplunL,ChungWC,RanaA,AtfiA,etal.Sirt2 deacetylaseisanovelAKTbindingpartnercriticalforAKTactivationby insulin.TheJournalofBiologicalChemistry2014;289:6054–66.
[76]RabadiMM,Xavier S,VaskoR, KaurK,GoligorksyMS,RatliffBB. High-mobilitygroupbox1isanoveldeacetylationtargetofSirtuin1.Kidney International2014.http://dx.doi.org/10.1038/ki.2014.217.
[77]WangRH,ZhengY,KimHS,XuX,CaoL,LuhasenT,etal.Interplayamong BRCA1,SIRT1,andSurvivinduringBRCA1-associatedtumorigenesis. Molec-ularCell2008;32:11–20.
[78]RothM,ChenWY.Sortingoutfunctionsofsirtuinsincancer.Oncogene 2014;33:1609–20.
[79]YaoY,YangY,ZhuWG.Sirtuins:nodesconnectingaging,metabolismand tumorigenesis.CurrentPharmaceuticalDesign2014;20:1614–24. [80]LuoJ,NikolaevAY,ImaiS,ChenD,SuF,ShilohA,etal.Negativecontrolofp53
bySir2alphapromotescellsurvivalunderstress.Cell2001;107:137–48. [81]VaziriH,DessainSK,NgEatonE,ImaiSI,FryeRA,PanditaTK,etal.hSIR2(SIRT1)
functionsasanNAD-dependentp53deacetylase.Cell2001;107:149–59. [82]VaqueroA,ScherM,LeeD,Erdjument-BromageH,TempstP,ReinbergD.
HumanSirT1interactswithhistoneH1andpromotesformationof faculta-tiveheterochromatin.MolecularCell2004;16:93–105.
[83]Bosch-PresegueL,Raurell-VilaH,Marazuela-DuqueA,Kane-GoldsmithN,Valle A,OliverJ,etal.StabilizationofSuv39H1bySirT1ispartofoxidativestress responseandensuresgenomeprotection.MolecularCell2011;42:210–23. [84]O‘HaganHM,MohammadHP,BaylinSB.Doublestrandbreakscaninitiate
genesilencingandSIRT1-dependentonsetofDNAmethylationinan exoge-nouspromoterCpGisland.PLoSGenetics2008;4:e1000155.
[85]Pruitt K,ZinnRL,Ohm JE,McGarvey KM,Kang SH,WatkinsDN,et al. InhibitionofSIRT1reactivatessilencedcancergeneswithoutlossof promot-erDNAhypermethylation.PLoSGenetics2006;2:e40.
[86]FanW,LuoJ.SIRT1regulatesUV-inducedDNArepairthroughdeacetylating XPA.MolecularCell2010;39:247–58.
[87]MingM,SheaCR,GuoX,LiX,SoltaniK,HanW,etal.Regulationofglobal genomenucleotideexcisionrepairbySIRT1throughxerodermapigmentosum C.ProceedingsoftheNationalAcademyofSciencesoftheUnitedStatesof America2010;107:22623–28.
[88]GaoY,FergusonDO,XieW,ManisJP,SekiguchiJ,FrankKM,etal.Interplayof p53andDNA-repairproteinXRCC4intumorigenesis,genomicstabilityand development.Nature2000;404:897–900.
[89]JeongJ,JuhnK,LeeH,KimSH,MinBH,LeeKM,etal.SIRT1promotesDNA repairactivityanddeacetylationofKu70.Experimental&Molecular Medi-cine2007;39:8–13.
[90]LiK,CastaA,WangR,LozadaE,FanW,KaneS,etal.RegulationofWRN proteincellular localizationandenzymaticactivities by SIRT1-mediated deacetylation.TheJournalofBiologicalChemistry2008;283:7590–8. [91]YuanZ,SetoE.AfunctionallinkbetweenSIRT1deacetylaseandNBS1inDNA
damageresponse.CellCycle2007;6:2869–71.
[92]YuanZ,ZhangX,SenguptaN,LaneWS,SetoE.SIRT1regulatesthefunctionof theNijmegenbreakagesyndromeprotein.MolecularCell2007;27:149–62. [93]WangRH,SenguptaK,LiC,KimHS,CaoL,XiaoC,etal.ImpairedDNAdamage response,genomeinstability, andtumorigenesis inSIRT1 mutantmice. CancerCell2008;14:312–23.
[94]OberdoerfferP,MichanS,McVayM,MostoslavskyR,VannJ,ParkSK,etal. SIRT1redistribution onchromatinpromotesgenomicstabilitybutalters geneexpressionduringaging.Cell2008;135:907–18.
[95]GuptaA,Guerin-PeyrouTG,SharmaGG,ParkC,AgarwalM,GanjuRK,etal. ThemammalianorthologofDrosophilaMOFthatacetylateshistoneH4 lysine16isessentialforembryogenesisandoncogenesis.Molecularand CellularBiology2008;28:397–409.
[96]ReaS,XouriG,AkhtarA.Malesabsentonthefirst(MOF):fromfliesto humans.Oncogene2007;26:5385–94.
[97]IkuraT,OgryzkoVV,GrigorievM,GroismanR,WangJ,HorikoshiM,etal. InvolvementoftheTIP60histone acetylasecomplexinDNArepair and apoptosis.Cell2000;102:463–73.
[98]PengL,LingH,YuanZ,FangB,BloomG,FukasawaK,etal.SIRT1negatively regulatestheactivities,functions,andproteinlevelsofhMOFandTIP60. MolecularandCellularBiology2012;32:2823–36.
[99]SunY,JiangX,ChenS,FernandesN,PriceBD.ArolefortheTip60histone acetyltransferaseintheacetylationandactivationofATM.Proceedingsofthe National Academy ofSciences of theUnitedStates ofAmerica2005;102:13182–87. [100]SawadaM,SunW,HayesP,LeskovK,BoothmanDA,MatsuyamaS.Ku70 suppressestheapoptotictranslocationofBaxtomitochondria.NatureCell Biology2003;5:320–9.
[101]CohenHY,LavuS,BittermanKJ,HekkingB,ImahiyeroboTA,MillerC,etal. AcetylationoftheC terminusofKu70by CBPandPCAFcontrols Bax-mediatedapoptosis.MolecularCell2004;13:627–38.
[102]CohenHY,MillerC,BittermanKJ,WallNR,HekkingB,KesslerB,etal.Calorie restrictionpromotesmammaliancellsurvivalbyinducingtheSIRT1 deace-tylase.Science2004;305:390–2.
[103]McBurneyMW,YangX,JardineK,BiemanM,Th’ngJ,LemieuxM.Theabsence ofSIR2alphaproteinhasnoeffectonglobalgenesilencinginmouse embry-onicstemcells.MolecularCancerResearch:MCR2003;1:402–9. [104]McBurneyMW,YangX,JardineK,HixonM,BoekelheideK,WebbJR,etal.The
mammalianSIR2alphaproteinhasaroleinembryogenesisand gametogen-esis.MolecularandCellularBiology2003;23:38–54.
[105]LiT,KonN,JiangL,TanM,LudwigT,ZhaoY,etal.Tumorsuppressioninthe absenceofp53-mediatedcell-cyclearrest,apoptosis,andsenescence.Cell 2012;149:1269–83.
[106]SuX,GiYJ,ChakravartiD,ChanIL,ZhangA,XiaX,etal.TAp63isamaster transcriptionalregulatoroflipidandglucosemetabolism.CellMetabolism 2012;16:511–25.
[107]RufiniA,Niklison-ChirouMV,InoueS,TomasiniR,HarrisIS,MarinoA,etal. TAp73depletionacceleratesagingthroughmetabolicdysregulation.Genes& Development2012;26:2009–14.
[108]BerkersCR,MaddocksOD,CheungEC,MorI,VousdenKH.Metabolic regula-tionbyp53familymembers.CellMetabolism2013;18:617–33.
[109]vonStechowL,Ruiz-AracamaA,vandeWaterB,PeijnenburgA,DanenE, Lommen A. Identification of cisplatin-regulated metabolic pathways in pluripotentstemcells.PLoSONE2013;8:e76476.
[110]LukasJ,LukasC,BartekJ.Morethanjustafocus:thechromatinresponseto DNAdamageanditsroleingenomeintegritymaintenance.NatureCell Biology2011;13:1161–9.
[111]ShiL,OberdoerfferP.ChromatindynamicsinDNAdouble-strandbreak repair.BiochimicaetBiophysicaActa2012;1819:811–9.
[112]SoriaG, PoloSE,Almouzni G. Prime, repair,restore: theactive roleof chromatinintheDNAdamageresponse.MolecularCell2012;46:722–34. [113]GonfloniS.TargetingDNAdamageresponse:threshold,chromatinlandscape
andbeyond.Pharmacology&Therapeutics2013;138:46–52.
[114]SmeenkG,vanAttikumH.ThechromatinresponsetoDNAbreaks:leavinga markongenomeintegrity.AnnualReviewofBiochemistry2013;82:55–80. [115]LiuJ,KimJ,OberdoerfferP.Metabolicmodulationofchromatin:implications
forDNArepairandgenomicintegrity.FrontiersinGenetics2013;4:182. [116]KruseJP,GuW.Modesofp53regulation.Cell2009;137:609–22. [117]ComelA,SorrentinoG,CapaciV,DelSalG.Thecytoplasmicsideofp53’s
oncosuppressive activities. FEBS Letters 2014;588(16):2600–9. http:// dx.doi.org/10.1016/j.febslet.2014.04.015.
[118]JiangP,DuW,YangX.p53andregulationoftumormetabolism.Journalof Carcinogenesis2013;12:21.
S.Gonflonietal./BiochemicalPharmacologyxxx(2014)xxx–xxx 8
GModel